A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos
- PMID: 19758566
- PMCID: PMC2746918
- DOI: 10.1016/j.devcel.2009.08.005
A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos
Abstract
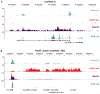
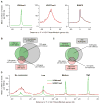
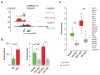
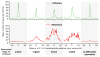
Epigenetic mechanisms set apart the active and inactive regions in the genome of multicellular organisms to produce distinct cell fates during embryogenesis. Here, we report on the epigenetic and transcriptome genome-wide maps of gastrula-stage Xenopus tropicalis embryos using massive parallel sequencing of cDNA (RNA-seq) and DNA obtained by chromatin immunoprecipitation (ChIP-seq) of histone H3 K4 and K27 trimethylation and RNA Polymerase II (RNAPII). These maps identify promoters and transcribed regions. Strikingly, genomic regions featuring opposing histone modifications are mostly transcribed, reflecting spatially regulated expression rather than bivalency as determined by expression profile analyses, sequential ChIP, and ChIP-seq on dissected embryos. Spatial differences in H3K27me3 deposition are predictive of localized gene expression. Moreover, the appearance of H3K4me3 coincides with zygotic gene activation, whereas H3K27me3 is predominantly deposited upon subsequent spatial restriction or repression of transcriptional regulators. These results reveal a hierarchy in the spatial control of zygotic gene activation.
Figures

Comment in
-
The curious case of bivalent marks.Dev Cell. 2009 Sep;17(3):301-3. doi: 10.1016/j.devcel.2009.08.014. Dev Cell. 2009. PMID: 19758552
References
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nature cell biology. 2006;8:532–538. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nature structural & molecular biology. 2007;14:1008–1016. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

