Cooperative transcriptional regulation of the essential pancreatic islet gene NeuroD1 (beta2) by Nkx2.2 and neurogenin 3
- PMID: 19759004
- PMCID: PMC2781522
- DOI: 10.1074/jbc.M109.048694
Cooperative transcriptional regulation of the essential pancreatic islet gene NeuroD1 (beta2) by Nkx2.2 and neurogenin 3
Abstract
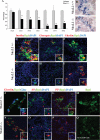
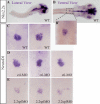
Nkx2.2 and NeuroD1 are two critical regulators of pancreatic beta cell development. Nkx2.2 is a homeodomain transcription factor that is essential for islet cell type specification and mature beta cell function. NeuroD1 is a basic helix-loop-helix transcription factor that is critical for islet beta cell maturation and maintenance. Although both proteins influence beta cell development directly downstream of the endocrine progenitor factor, neurogenin3 (Ngn3), a connection between the two proteins in the regulation of beta cell fate and function has yet to be established. In this study, we demonstrate that Nkx2.2 transcriptional activity is required to facilitate the activation of NeuroD1 by Ngn3. Furthermore, Nkx2.2 is necessary to maintain high levels of NeuroD1 expression in developing mouse and zebrafish islets and in mature beta cells. Interestingly, Nkx2.2 regulates NeuroD1 through two independent promoter elements, one that is bound and activated directly by Nkx2.2 and one that appears to be regulated by Nkx2.2 through an indirect mechanism. Together, these findings suggest that Nkx2.2 coordinately activates NeuroD1 with Ngn3 within the endocrine progenitor cell and also plays a role in the maintenance of NeuroD1 expression to regulate beta cell function in the mature islet. Collectively, these findings further define the conserved regulatory networks involved in islet beta cell formation and function.
Figures




References
-
- Pictet R., Rutter W. J. (1972) Development of Embryonic Endocrine Pancreas, pp. 26–28, Williams & Wilkins, Washington, DC
-
- Murtaugh L. C. (2007) Development 134, 427–438 - PubMed
-
- Jørgensen M. C., Ahnfelt-Rønne J., Hald J., Madsen O. D., Serup P., Hecksher-Sørensen J. (2007) Endocr. Rev. 28, 685–705 - PubMed
-
- Collombat P., Hecksher-Sørensen J., Serup P., Mansouri A. (2006) Mech. dev. 123, 501–512 - PubMed
-
- Habener J. F., Kemp D. M., Thomas M. K. (2005) Endocrinology 146, 1025–1034 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

