SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression
- PMID: 19759026
- PMCID: PMC2781707
- DOI: 10.1074/jbc.M109.058610
SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression
Abstract
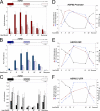
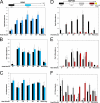
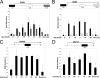
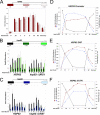
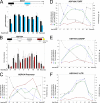
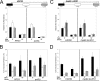
The chromatin structure of heat shock protein (HSP)-encoding genes undergoes dramatic alterations upon transcriptional induction, including, in extreme cases, domain-wide nucleosome disassembly. Here, we use a combination of gene knock-out, in situ mutagenesis, chromatin immunoprecipitation, and expression assays to investigate the role of histone modification complexes in regulating heat shock gene structure and expression in Saccharomyces cerevisiae. Two histone acetyltransferases, Gcn5 and Esa1, were found to stimulate HSP gene transcription. A detailed chromatin immunoprecipitation analysis of the Gcn5-containing SAGA complex (signified by Spt3) revealed its presence within the promoter of every heat shock factor 1-regulated gene examined. The occupancy of SAGA increased substantially upon heat shock, peaking at several HSP promoters within 30-45 s of temperature upshift. SAGA was also efficiently recruited to the coding regions of certain HSP genes (where its presence mirrored that of pol II), although not at others. Robust and rapid recruitment of repressive, Rpd3-containing histone deacetylase complexes was also seen and at all HSP genes examined. A detailed analysis of HSP82 revealed that both Rpd3(L) and Rpd3(S) complexes (signified by Sap30 and Rco1, respectively) were recruited to the gene promoter, yet only Rpd3(S) was recruited to its open reading frame. A consensus URS1 cis-element facilitated the recruitment of each Rpd3 complex to the HSP82 promoter, and this correlated with targeted deacetylation of promoter nucleosomes. Collectively, our observations reveal that SAGA and Rpd3 complexes are rapidly and synchronously recruited to heat shock factor 1-activated genes and suggest that their opposing activities modulate heat shock gene chromatin structure and fine-tune transcriptional output.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

