Transcription from bacteriophage lambda pR promoter is regulated independently and antagonistically by DksA and ppGpp
- PMID: 19759216
- PMCID: PMC2777414
- DOI: 10.1093/nar/gkp676
Transcription from bacteriophage lambda pR promoter is regulated independently and antagonistically by DksA and ppGpp
Abstract
The stringent response effector, guanosine tetraphosphate (ppGpp), adjust gene expression and physiology in bacteria, by affecting the activity of various promoters. RNA polymerase-interacting protein, DksA, was proposed to be the co-factor of ppGpp effects; however, there are reports suggesting independent roles of these regulators. Bacteriophage lambda major lytic promoter, pR, is down-regulated by the stringent response and ppGpp. Here, we present evidence that DksA significantly stimulates pR-initiated transcription in vitro in the reconstituted system. DksA is also indispensable for pR activity in vivo. DksA-mediated activation of pR-initiated transcription is predominant over ppGpp effects in the presence of both regulators in vitro. The possible role of the opposite regulation by ppGpp and DksA in lambda phage development is discussed. The major mechanism of DksA-mediated activation of transcription from pR involves facilitating of RNA polymerase binding to the promoter region, which results in more productive transcription initiation. Thus, our results provide evidence for the first promoter inhibited by ppGpp that can be stimulated by the DksA protein both in vivo and in vitro. Therefore, DksA role could be not only independent but antagonistic to ppGpp in transcription regulation.
Figures
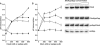


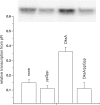

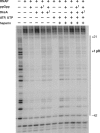
References
-
- Cashel M, Gentry D, Hernandez VJ, Vinella D. Escherichia coli and Salmonella: Cellular and Molecular Biology. I. Washington DC: American Society for Microbiology; 1996. The stringent response; pp. 1458–1496.
-
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. - PubMed
-
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. - PubMed
-
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. - PubMed
-
- Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Vassylyev DG, Ross W, Gourse RL. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 2008;377:551–564. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

