Novel circular DNA viruses in stool samples of wild-living chimpanzees
- PMID: 19759238
- PMCID: PMC2887567
- DOI: 10.1099/vir.0.015446-0
Novel circular DNA viruses in stool samples of wild-living chimpanzees
Abstract
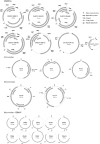
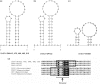
Viral particles in stool samples from wild-living chimpanzees were analysed using random PCR amplification and sequencing. Sequences encoding proteins distantly related to the replicase protein of single-stranded circular DNA viruses were identified. Inverse PCR was used to amplify and sequence multiple small circular DNA viral genomes. The viral genomes were related in size and genome organization to vertebrate circoviruses and plant geminiviruses but with a different location for the stem-loop structure involved in rolling circle DNA replication. The replicase genes of these viruses were most closely related to those of the much smaller (approximately 1 kb) plant nanovirus circular DNA chromosomes. Because the viruses have characteristics of both animal and plant viruses, we named them chimpanzee stool-associated circular viruses (ChiSCV). Further metagenomic studies of animal samples will greatly increase our knowledge of viral diversity and evolution.
Figures



References
-
- Bassami, M. R., Berryman, D., Wilcox, G. E. & Raidal, S. R. (1998). Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anaemia virus. Virology 249, 453–459. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

