Design parameters to control synthetic gene expression in Escherichia coli
- PMID: 19759823
- PMCID: PMC2736378
- DOI: 10.1371/journal.pone.0007002
Design parameters to control synthetic gene expression in Escherichia coli
Abstract
Background: Production of proteins as therapeutic agents, research reagents and molecular tools frequently depends on expression in heterologous hosts. Synthetic genes are increasingly used for protein production because sequence information is easier to obtain than the corresponding physical DNA. Protein-coding sequences are commonly re-designed to enhance expression, but there are no experimentally supported design principles.
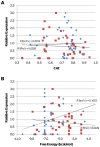
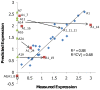
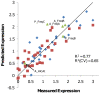
Principal findings: To identify sequence features that affect protein expression we synthesized and expressed in E. coli two sets of 40 genes encoding two commercially valuable proteins, a DNA polymerase and a single chain antibody. Genes differing only in synonymous codon usage expressed protein at levels ranging from undetectable to 30% of cellular protein. Using partial least squares regression we tested the correlation of protein production levels with parameters that have been reported to affect expression. We found that the amount of protein produced in E. coli was strongly dependent on the codons used to encode a subset of amino acids. Favorable codons were predominantly those read by tRNAs that are most highly charged during amino acid starvation, not codons that are most abundant in highly expressed E. coli proteins. Finally we confirmed the validity of our models by designing, synthesizing and testing new genes using codon biases predicted to perform well.
Conclusion: The systematic analysis of gene design parameters shown in this study has allowed us to identify codon usage within a gene as a critical determinant of achievable protein expression levels in E. coli. We propose a biochemical basis for this, as well as design algorithms to ensure high protein production from synthetic genes. Replication of this methodology should allow similar design algorithms to be empirically derived for any expression system.
Conflict of interest statement
Figures



References
-
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. - PubMed
-
- Newcomb J, Carlson R, Aldrich SC. Cambridge, MA: Bio Economic Research Associates; 2007. Genome synthesis and design futures: Implications for the U.S. economy.
-
- Itakura K, Hirose T, Crea R, Riggs AD, Heyneker HL, et al. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1977;198:1056–1063. - PubMed
-
- Henaut A, Danchin A. Analysis and predictions from Escherichia coli sequences. In: Neidhardt FC, Curtiss RI, Ingraham J, Lin E, Brooks Low K, et al., editors. Escherichia coli and Salmonella typhimurium cellular and molecular biology. Washington, D.C: ASM press; 1996. pp. 2047–2066.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

