Transportin mediates nuclear entry of DNA in vertebrate systems
- PMID: 19761539
- PMCID: PMC3073982
- DOI: 10.1111/j.1600-0854.2009.00968.x
Transportin mediates nuclear entry of DNA in vertebrate systems
Abstract
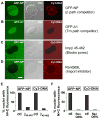
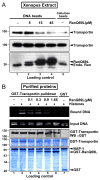
Delivery of DNA to the cell nucleus is an essential step in many types of viral infection, transfection, gene transfer by the plant pathogen Agrobacterium tumefaciens and in strategies for gene therapy. Thus, the mechanism by which DNA crosses the nuclear pore complex (NPC) is of great interest. Using nuclei reconstituted in vitro in Xenopus egg extracts, we previously studied DNA passage through the nuclear pores using a single-molecule approach based on optical tweezers. Fluorescently labeled DNA molecules were also seen to accumulate within nuclei. Here we find that this import of DNA relies on a soluble protein receptor of the importin family. To identify this receptor, we used different pathway-specific cargoes in competition studies as well as pathway-specific dominant negative inhibitors derived from the nucleoporin Nup153. We found that inhibition of the receptor transportin suppresses DNA import. In contrast, inhibition of importin beta has little effect on the nuclear accumulation of DNA. The dependence on transportin was fully confirmed in assays using permeabilized HeLa cells and a mammalian cell extract. We conclude that the nuclear import of DNA observed in these different vertebrate systems is largely mediated by the receptor transportin. We further report that histones, a known cargo of transportin, can act as an adaptor for the binding of transportin to DNA.
Figures





Similar articles
-
Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors.Curr Biol. 1998 Dec 17-31;8(25):1376-86. doi: 10.1016/s0960-9822(98)00018-9. Curr Biol. 1998. PMID: 9889100
-
A role for transportin in the nuclear import of adenovirus core proteins and DNA.Traffic. 2007 Oct;8(10):1313-22. doi: 10.1111/j.1600-0854.2007.00618.x. Traffic. 2007. PMID: 17848175 Free PMC article.
-
Transportin acts to regulate mitotic assembly events by target binding rather than Ran sequestration.Mol Biol Cell. 2014 Apr;25(7):992-1009. doi: 10.1091/mbc.E13-08-0506. Epub 2014 Jan 29. Mol Biol Cell. 2014. PMID: 24478460 Free PMC article.
-
Structural Biology and Regulation of Protein Import into the Nucleus.J Mol Biol. 2016 May 22;428(10 Pt A):2060-90. doi: 10.1016/j.jmb.2015.10.023. Epub 2015 Oct 30. J Mol Biol. 2016. PMID: 26523678 Review.
-
The nuclear pore complex: the gateway to successful nonviral gene delivery.Pharm Res. 2006 Mar;23(3):447-59. doi: 10.1007/s11095-005-9445-4. Epub 2006 Mar 15. Pharm Res. 2006. PMID: 16525863 Review.
Cited by
-
Uncoupling uncoating of herpes simplex virus genomes from their nuclear import and gene expression.J Virol. 2011 May;85(9):4271-83. doi: 10.1128/JVI.02067-10. Epub 2011 Feb 23. J Virol. 2011. PMID: 21345968 Free PMC article.
-
Analysis of nuclear reconstitution, nuclear envelope assembly, and nuclear pore assembly using Xenopus in vitro assays.Methods Cell Biol. 2014;122:165-91. doi: 10.1016/B978-0-12-417160-2.00008-4. Methods Cell Biol. 2014. PMID: 24857730 Free PMC article.
-
Nanoparticle-based technologies for retinal gene therapy.Eur J Pharm Biopharm. 2015 Sep;95(Pt B):353-67. doi: 10.1016/j.ejpb.2014.12.028. Epub 2015 Jan 12. Eur J Pharm Biopharm. 2015. PMID: 25592325 Free PMC article. Review.
-
Mechanisms of unprimed and dexamethasone-primed nonviral gene delivery to human mesenchymal stem cells.Biotechnol Bioeng. 2019 Feb;116(2):427-443. doi: 10.1002/bit.26870. Epub 2018 Dec 7. Biotechnol Bioeng. 2019. PMID: 30450542 Free PMC article.
-
In vitro assembly of nuclear envelope in tobacco cultured cells.Nucleus. 2021 Dec;12(1):82-89. doi: 10.1080/19491034.2021.1930681. Nucleus. 2021. PMID: 34030583 Free PMC article.
References
-
- Kasamatsu H, Nakanishi A. How do animal DNA viruses get to the nucleus? Annu Rev Microbiol. 1998;52:627–86. - PubMed
-
- Lechardeur D, Lukacs GL. Nucleocytoplasmic transport of plasmid DNA: a perilous journey from the cytoplasm to the nucleus. Hum Gene Ther. 2006;17:882–9. - PubMed
-
- Pouton CW, Wagstaff KM, Roth DM, Moseley GW, Jans DA. Targeted delivery to the nucleus. Adv Drug Deliv Rev. 2007;59:698–717. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

