Simultaneous alignment of short reads against multiple genomes
- PMID: 19761611
- PMCID: PMC2768987
- DOI: 10.1186/gb-2009-10-9-r98
Simultaneous alignment of short reads against multiple genomes
Abstract
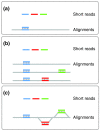
Genome resequencing with short reads generally relies on alignments against a single reference. GenomeMapper supports simultaneous mapping of short reads against multiple genomes by integrating related genomes (e.g., individuals of the same species) into a single graph structure. It constitutes the first approach for handling multiple references and introduces representations for alignments against complex structures. Demonstrated benefits include access to polymorphisms that cannot be identified by alignments against the reference alone. Download GenomeMapper at http://1001genomes.org.
Figures


References
-
- Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, Magrini VJ, Richt RJ, Sander SN, Stewart DA, Stromberg M, Tsung EF, Wylie T, Schedl T, Wilson RK, Mardis ER. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. - DOI - PubMed
-
- Ahn SM, Kim TH, Lee S, Kim D, Ghang H, Kim DS, Kim BC, Kim SY, Kim WY, Kim C, Park D, Lee YS, Kim S, Reja R, Jho S, Kim CG, Cha JY, Kim KH, Lee B, Bhak J, Kim SJ. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. - DOI - PMC - PubMed
-
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. - DOI - PMC - PubMed
-
- Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, Kim HS, Chung IS, Lee WC, Lee JS, Seo SH, Yun JY, Woo HN, Lee H, Suh D, Kim HJ, Yavartanoo M, Kwak M, Zheng Y, Lee MK, Kim JY, Gokcumen O, Mills RE, Zaranek AW, Thakuria J, Wu X, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

