An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons
- PMID: 19762510
- PMCID: PMC2758739
- DOI: 10.1101/gad.1837009
An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons
Abstract
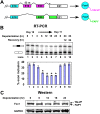
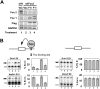
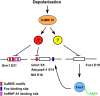
Neuronal depolarization and CaM kinase IV signaling alter the splicing of multiple exons in transcripts for ion channels, neurotransmitter receptors, and other synaptic proteins. These splicing changes are mediated in part by special CaM kinase-responsive RNA elements, within or adjacent to exons that are repressed in the initial phase of chronic depolarization. The splicing of many neuronal transcripts is also regulated by members of the Fox (Feminizing gene on X) protein family, and these Fox targets are also often proteins affecting synaptic activity. We show that Fox-1/Ataxin 2-Binding Protein 1 (A2BP1), a protein implicated in a variety of neurological diseases, can counteract the effects of chronic depolarization on splicing. We find that exon 19 of Fox-1 is itself repressed by depolarization. Fox-1 transcripts missing exon 19 encode a nuclear isoform of Fox-1 that progressively replaces the cytoplasmic Fox-1 isoform as cells are maintained depolarizing media. The resulting increase in nuclear Fox-1 leads to the reactivation of many Fox-1 target exons, including exon 5 of the NMDA receptor 1, that were initially repressed by the high-KCl medium. These results reveal a novel mechanism for the slow modulation of splicing as cells adapt to chronic stimuli: The subcellular localization of a splicing regulator is controlled through its own alternative splicing.
Figures


References
-
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with image J. Biophotonics Int. 2004;11:36–42.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
