A longevity protein, Lag2, interacts with SCF complex and regulates SCF function
- PMID: 19763088
- PMCID: PMC2776101
- DOI: 10.1038/emboj.2009.268
A longevity protein, Lag2, interacts with SCF complex and regulates SCF function
Abstract
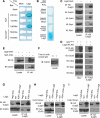
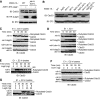
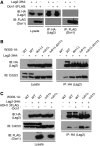
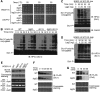
SCF-type E3-ubiquitin ligases control numerous cellular processes through the ubiquitin-proteasome pathway. However, the regulation of SCF function remains largely uncharacterized. Here, we report a novel SCF complex-interacting protein, Lag2, in Saccharomyces cerevisiae. Lag2 interacts with the SCF complex under physiological conditions. Lag2 negatively controls the ubiquitylation activities of SCF E3 ligase by interrupting the association of Cdc34 to SCF complex. Overexpression of Lag2 increases unrubylated Cdc53, whereas deletion of lag2, together with the deletions of dcn1 and jab1, results in the accumulation of Rub1-modified Cdc53. In vitro rubylation assays show that Lag2 inhibits the conjugation of Rub1 to Cdc53 in competition with Dcn1, which suggest that Lag2 down-regulates the rubylation of Cdc53 rather than promoting derubylation. Furthermore, Dcn1 hinders the association of Lag2 to Cdc53 in vivo. Finally, the deletion of lag2 combined with the deletion of either dcn1 or rub1 suppresses the growth of yeast cells. These observations thus indicate that Lag2 has a significant function in regulating the SCF complex by controlling its ubiquitin ligase activities and its rubylation cycle.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae.EMBO J. 2009 Dec 16;28(24):3845-56. doi: 10.1038/emboj.2009.354. EMBO J. 2009. PMID: 19942853 Free PMC article.
-
A dual E3 mechanism for Rub1 ligation to Cdc53.Mol Cell. 2010 Sep 10;39(5):784-96. doi: 10.1016/j.molcel.2010.08.030. Mol Cell. 2010. PMID: 20832729 Free PMC article.
-
The Cdc34/SCF ubiquitination complex mediates Saccharomyces cerevisiae cell wall integrity.Genetics. 2006 Dec;174(4):1825-39. doi: 10.1534/genetics.106.059154. Epub 2006 Oct 8. Genetics. 2006. PMID: 17028344 Free PMC article.
-
Proteolytic regulation of metabolic enzymes by E3 ubiquitin ligase complexes: lessons from yeast.Crit Rev Biochem Mol Biol. 2015;50(6):489-502. doi: 10.3109/10409238.2015.1081869. Epub 2015 Sep 11. Crit Rev Biochem Mol Biol. 2015. PMID: 26362128 Review.
-
The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction.Prog Biophys Mol Biol. 1999;72(3):299-328. doi: 10.1016/s0079-6107(99)00010-3. Prog Biophys Mol Biol. 1999. PMID: 10581972 Review.
Cited by
-
Cand1-Mediated Adaptive Exchange Mechanism Enables Variation in F-Box Protein Expression.Mol Cell. 2018 Mar 1;69(5):773-786.e6. doi: 10.1016/j.molcel.2018.01.038. Mol Cell. 2018. PMID: 29499133 Free PMC article.
-
New insight into the role of the Cdc34 ubiquitin-conjugating enzyme in cell cycle regulation via Ace2 and Sic1.Genetics. 2011 Mar;187(3):701-15. doi: 10.1534/genetics.110.125302. Epub 2010 Dec 31. Genetics. 2011. PMID: 21196523 Free PMC article.
-
CSN- and CAND1-dependent remodelling of the budding yeast SCF complex.Nat Commun. 2013;4:1641. doi: 10.1038/ncomms2628. Nat Commun. 2013. PMID: 23535662
-
Structural and functional insights to ubiquitin-like protein conjugation.Annu Rev Biophys. 2014;43:357-79. doi: 10.1146/annurev-biophys-051013-022958. Annu Rev Biophys. 2014. PMID: 24773014 Free PMC article. Review.
-
Hermes Transposon Mutagenesis Shows [URE3] Prion Pathology Prevented by a Ubiquitin-Targeting Protein: Evidence for Carbon/Nitrogen Assimilation Cross Talk and a Second Function for Ure2p in Saccharomyces cerevisiae.Genetics. 2018 Jul;209(3):789-800. doi: 10.1534/genetics.118.300981. Epub 2018 May 16. Genetics. 2018. PMID: 29769283 Free PMC article.
References
-
- Avaro S, Belgareh-Touze N, Sibella-Arguelles C, Volland C, Haguenauer-Tsapis R (2002) Mutants defective in secretory/vacuolar pathways in the EUROFAN collection of yeast disruptants. Yeast 19: 351–371 - PubMed
-
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 - PubMed
-
- Childress AM, Franklin DS, Pinswasdi C, Kale S (1996) LAG2, a gene that determines yeast longevity. Microbiology 142 (Part 8): 2289–2297 - PubMed
-
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

