A role of SCN9A in human epilepsies, as a cause of febrile seizures and as a potential modifier of Dravet syndrome
- PMID: 19763161
- PMCID: PMC2730533
- DOI: 10.1371/journal.pgen.1000649
A role of SCN9A in human epilepsies, as a cause of febrile seizures and as a potential modifier of Dravet syndrome
Abstract
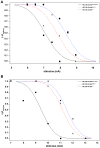
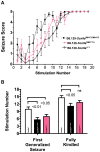
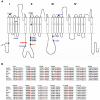
A follow-up study of a large Utah family with significant linkage to chromosome 2q24 led us to identify a new febrile seizure (FS) gene, SCN9A encoding Na(v)1.7. In 21 affected members, we uncovered a potential mutation in a highly conserved amino acid, p.N641Y, in the large cytoplasmic loop between transmembrane domains I and II that was absent from 586 ethnically matched population control chromosomes. To establish a functional role for this mutation in seizure susceptibility, we introduced the orthologous mutation into the murine Scn9a ortholog using targeted homologous recombination. Compared to wild-type mice, homozygous Scn9a(N641Y/N641Y) knockin mice exhibit significantly reduced thresholds to electrically induced clonic and tonic-clonic seizures, and increased corneal kindling acquisition rates. Together, these data strongly support the SCN9A p.N641Y mutation as disease-causing in this family. To confirm the role of SCN9A in FS, we analyzed a collection of 92 unrelated FS patients and identified additional highly conserved Na(v)1.7 missense variants in 5% of the patients. After one of these children with FS later developed Dravet syndrome (severe myoclonic epilepsy of infancy), we sequenced the SCN1A gene, a gene known to be associated with Dravet syndrome, and identified a heterozygous frameshift mutation. Subsequent analysis of 109 Dravet syndrome patients yielded nine Na(v)1.7 missense variants (8% of the patients), all in highly conserved amino acids. Six of these Dravet syndrome patients with SCN9A missense variants also harbored either missense or splice site SCN1A mutations and three had no SCN1A mutations. This study provides evidence for a role of SCN9A in human epilepsies, both as a cause of FS and as a partner with SCN1A mutations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Comment in
-
SCN9A: another sodium channel excited to play a role in human epilepsies.Clin Genet. 2010 Apr;77(4):326-8. doi: 10.1111/j.1399-0004.2009.01366_1.x. Epub 2010 Jan 20. Clin Genet. 2010. PMID: 20095983 No abstract available.
References
-
- Arzimanoglou A, Guerrini R, Aicardi J. Aicardi's Epilepsy in Children. 3rd ed. New York: Lippincott Williams & Wilkins; 2004. pp. 51–57.
-
- Singh R, Scheffer IE, Crossland K, Berkovic SF. Generalized Epilepsy with Febrile Seizures Plus: A Common Childhood-Onset Genetic Epilepsy Syndrome. Annals of Neurology. 1999;45:75–81. - PubMed
-
- Racacho LJ, McLachlan RS, Ebers GC, Maher J, Bulman DE. Evidence favoring genetic heterogeneity for febrile convulsions. Epilepsia. 2000;41:132–139. - PubMed
-
- Claes L, Ceulemans B, Audenaert D, Smets K, Lofgren A, et al. De novo SCN1A mutations are a major cause of severe myoclonic epilepsy of infancy. Hum Mutat. 2003;21:615–621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

