Finding local communities in protein networks
- PMID: 19765306
- PMCID: PMC2755114
- DOI: 10.1186/1471-2105-10-297
Finding local communities in protein networks
Abstract
Background: Protein-protein interactions (PPIs) play fundamental roles in nearly all biological processes, and provide major insights into the inner workings of cells. A vast amount of PPI data for various organisms is available from BioGRID and other sources. The identification of communities in PPI networks is of great interest because they often reveal previously unknown functional ties between proteins. A large number of global clustering algorithms have been applied to protein networks, where the entire network is partitioned into clusters. Here we take a different approach by looking for local communities in PPI networks.
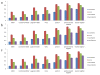
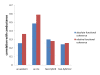
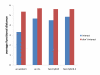
Results: We develop a tool, named Local Protein Community Finder, which quickly finds a community close to a queried protein in any network available from BioGRID or specified by the user. Our tool uses two new local clustering algorithms Nibble and PageRank-Nibble, which look for a good cluster among the most popular destinations of a short random walk from the queried vertex. The quality of a cluster is determined by proportion of outgoing edges, known as conductance, which is a relative measure particularly useful in undersampled networks. We show that the two local clustering algorithms find communities that not only form excellent clusters, but are also likely to be biologically relevant functional components. We compare the performance of Nibble and PageRank-Nibble to other popular and effective graph partitioning algorithms, and show that they find better clusters in the graph. Moreover, Nibble and PageRank-Nibble find communities that are more functionally coherent.
Conclusion: The Local Protein Community Finder, accessible at http://xialab.bu.edu/resources/lpcf, allows the user to quickly find a high-quality community close to a queried protein in any network available from BioGRID or specified by the user. We show that the communities found by our tool form good clusters and are functionally coherent, making our application useful for biologists who wish to investigate functional modules that a particular protein is a part of.
Figures

Similar articles
-
Spectral affinity in protein networks.BMC Syst Biol. 2009 Nov 29;3:112. doi: 10.1186/1752-0509-3-112. BMC Syst Biol. 2009. PMID: 19943959 Free PMC article.
-
Identification of Protein Complexes Using Weighted PageRank-Nibble Algorithm and Core-Attachment Structure.IEEE/ACM Trans Comput Biol Bioinform. 2015 Jan-Feb;12(1):179-92. doi: 10.1109/TCBB.2014.2343954. IEEE/ACM Trans Comput Biol Bioinform. 2015. PMID: 26357088
-
IsoRankN: spectral methods for global alignment of multiple protein networks.Bioinformatics. 2009 Jun 15;25(12):i253-8. doi: 10.1093/bioinformatics/btp203. Bioinformatics. 2009. PMID: 19477996 Free PMC article.
-
The function of communities in protein interaction networks at multiple scales.BMC Syst Biol. 2010 Jul 22;4:100. doi: 10.1186/1752-0509-4-100. BMC Syst Biol. 2010. PMID: 20649971 Free PMC article.
-
Some perspectives on network modeling in therapeutic target prediction.Biomed Eng Comput Biol. 2013 Feb 21;5:17-24. doi: 10.4137/BECB.S10793. eCollection 2013. Biomed Eng Comput Biol. 2013. PMID: 25288898 Free PMC article. Review.
Cited by
-
CompNet: a GUI based tool for comparison of multiple biological interaction networks.BMC Bioinformatics. 2016 Apr 26;17(1):185. doi: 10.1186/s12859-016-1013-x. BMC Bioinformatics. 2016. PMID: 27112575 Free PMC article.
-
Community structure detection for overlapping modules through mathematical programming in protein interaction networks.PLoS One. 2014 Nov 20;9(11):e112821. doi: 10.1371/journal.pone.0112821. eCollection 2014. PLoS One. 2014. PMID: 25412367 Free PMC article.
-
Distinctive Behaviors of Druggable Proteins in Cellular Networks.PLoS Comput Biol. 2015 Dec 23;11(12):e1004597. doi: 10.1371/journal.pcbi.1004597. eCollection 2015 Dec. PLoS Comput Biol. 2015. PMID: 26699810 Free PMC article.
-
SurvNet: a web server for identifying network-based biomarkers that most correlate with patient survival data.Nucleic Acids Res. 2012 Jul;40(Web Server issue):W123-6. doi: 10.1093/nar/gks386. Epub 2012 May 8. Nucleic Acids Res. 2012. PMID: 22570412 Free PMC article.
-
Heterodimeric protein complex identification by naïve Bayes classifiers.BMC Bioinformatics. 2013 Dec 3;14:347. doi: 10.1186/1471-2105-14-347. BMC Bioinformatics. 2013. PMID: 24299017 Free PMC article.
References
-
- Krogan N, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis A, Punna T, Peregrin-Alvarez J, Shales M, Zhang X, Davey M, Robinson M, Paccanaro A, Bray J, Sheung A, Beattie B, Richards D, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete M, Vlasblom J, Wu S, Orsi C, Collins S, Chandran S, Haw R, Rilstone J, Gandi K, Thompson N, Musso G, Onge PS, Ghanny S, Lam M, Butland G, Altaf-U A, Kanaya S, Shilatifard A, O'Shea E, Weissman J, Ingles J, Hughes T, Parkinson J, Gerstein M, Wodak S, Emili A, Greenblatt J. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. - DOI - PubMed
-
- Clauset A. Finding local community structure in networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;72:026132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

