The completion of the Mammalian Gene Collection (MGC)
- PMID: 19767417
- PMCID: PMC2792178
- DOI: 10.1101/gr.095976.109
The completion of the Mammalian Gene Collection (MGC)
Abstract
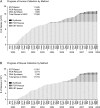
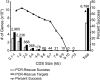
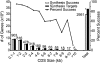
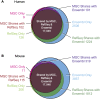
Since its start, the Mammalian Gene Collection (MGC) has sought to provide at least one full-protein-coding sequence cDNA clone for every human and mouse gene with a RefSeq transcript, and at least 6200 rat genes. The MGC cloning effort initially relied on random expressed sequence tag screening of cDNA libraries. Here, we summarize our recent progress using directed RT-PCR cloning and DNA synthesis. The MGC now contains clones with the entire protein-coding sequence for 92% of human and 89% of mouse genes with curated RefSeq (NM-accession) transcripts, and for 97% of human and 96% of mouse genes with curated RefSeq transcripts that have one or more PubMed publications, in addition to clones for more than 6300 rat genes. These high-quality MGC clones and their sequences are accessible without restriction to researchers worldwide.
Figures

References
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
