Complex architecture and regulated expression of the Sox2ot locus during vertebrate development
- PMID: 19767420
- PMCID: PMC2764477
- DOI: 10.1261/rna.1705309
Complex architecture and regulated expression of the Sox2ot locus during vertebrate development
Abstract
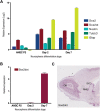
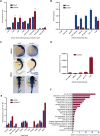
The Sox2 gene is a key regulator of pluripotency embedded within an intron of a long noncoding RNA (ncRNA), termed Sox2 overlapping transcript (Sox2ot), which is transcribed in the same orientation. However, this ncRNA remains uncharacterized. Here we show that Sox2ot has multiple transcription start sites associated with genomic features that indicate regulated expression, including highly conserved elements (HCEs) and chromatin marks characteristic of gene promoters. To identify biological processes in which Sox2ot may be involved, we analyzed its expression in several developmental systems, compared to expression of Sox2. We show that Sox2ot is a stable transcript expressed in mouse embryonic stem cells, which, like Sox2, is down-regulated upon induction of embryoid body (EB) differentiation. However, in contrast to Sox2, Sox2ot is up-regulated during EB mesoderm-lineage differentiation. In adult mouse, Sox2ot isoforms were detected in tissues where Sox2 is expressed, as well as in different tissues, supporting independent regulation of expression of the ncRNA. Sox2dot, an isoform of Sox2ot transcribed from a distal HCE located >500 kb upstream of Sox2, was detected exclusively in the mouse brain, with enrichment in regions of adult neurogenesis. In addition, Sox2ot isoforms are transcribed from HCEs upstream of Sox2 in other vertebrates, including in several regions of the human brain. We also show that Sox2ot is dynamically regulated during chicken and zebrafish embryogenesis, consistently associated with central nervous system structures. These observations provide insight into the structure and regulation of the Sox2ot gene, and suggest conserved roles for Sox2ot orthologs during vertebrate development.
Figures


References
-
- Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. - PubMed
-
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. - PubMed
-
- Andrew T, Maniatis N, Carbonaro F, Liew SH, Lau W, Spector TD, Hammond CJ. Identification and replication of three novel myopia common susceptibility gene loci on chromosome 3q26 using linkage and linkage disequilibrium mapping. PLoS Genet. 2008;4:e1000220. doi: 10.1371/journal.pgen.1000220. - DOI - PMC - PubMed
-
- Badger JH, Olsen GJ. CRITICA: Coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;16:512–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
