A proteomics approach to discovering natural products and their biosynthetic pathways
- PMID: 19767731
- PMCID: PMC2782881
- DOI: 10.1038/nbt.1565
A proteomics approach to discovering natural products and their biosynthetic pathways
Abstract
Many natural products with antibiotic, anticancer and antifungal properties are synthesized by nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs). Although genome sequencing has revealed the diversity of these enzymes, identifying new products and their biosynthetic pathways remains challenging. By taking advantage of the size of these enzymes (often >2,000 amino acids) and unique marker ions derived from their common phosphopantetheinyl cofactor, we adapted mass spectrometry-based proteomics to selectively detect NRPS and PKS gene clusters in microbial proteomes without requiring genome sequence information. We detected known NRPS systems in members of the genera Bacillus and Streptomyces, and screened 22 environmental isolates to uncover production of unknown natural products from the hybrid NRPS-PKS zwittermicin A biosynthetic gene cluster. We also discovered an NRPS cluster that generates a seven-residue lipopeptide. This 'protein-first' strategy complements bioassay- and sequence-based approaches by finding expressed gene clusters that produce new natural products.
Figures


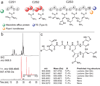
References
-
- Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. - PubMed
-
- Zerikly M, Challis GL. Strategies for the discovery of new natural products by genome mining. Chembiochem. 2009;10:625–633. - PubMed
-
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. - PubMed
-
- Weinstein MJ, Wagman GH. Antibiotics: isolation, separation, and purification. New York: Elsevier Scientific Publishing Company, New York; 1978.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

