Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1)
- PMID: 19768134
- PMCID: PMC2746644
- DOI: 10.1111/j.1750-2659.2009.00096.x
Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1)
Abstract
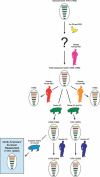
The novel H1N1 influenza virus that emerged in humans in Mexico in early 2009 and transmitted efficiently in the human population with global spread has been declared a pandemic strain. Here we review influenza infections in swine since 1918 and the introduction of different avian and human influenza virus genes into swine influenza viruses of North America and Eurasia. These introductions often result in viruses of increased fitness for pigs that occasionally transmit to humans. The novel virus affecting humans is derived from a North American swine influenza virus that has acquired two gene segments [Neuraminidase (NA) and Matrix (M)] from the European swine lineages. This reassortant appears to have increased fitness in humans. The potential for increased virulence in humans and of further reassortment between the novel H1N1 influenza virus and oseltamivir resistant seasonal H1N1 or with highly pathogenic H5N1 influenza stresses the need for urgent pandemic planning.
Figures
References
-
- Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virol 1983; 127:361–373. - PubMed
-
- Scholtissek C. Pigs as the “mixing vessel” for the creation of new pandemic influenza A viruses. Med Principles Pract 1990; 2:65–71.
-
- Koen JS. A practical method for field diagnosis of swine diseases. Am J Vet Med 1919; 14:468–470.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

