New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia
- PMID: 19770377
- PMCID: PMC2773477
- DOI: 10.1200/JCO.2009.21.9840
New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia
Abstract
Purpose: Cytogenetics is the primary outcome predictor in acute myeloid leukemia (AML). Metaphase cytogenetics (MC) detects an abnormal karyotype in only half of patients with AML, however. Single nucleotide polymorphism arrays (SNP-A) can detect acquired somatic uniparental disomy (UPD) and other cryptic defects, even in samples deemed normal by MC. We hypothesized that SNP-A will improve detection of chromosomal defects in AML and that this would enhance the prognostic value of MC.
Patients and methods: We performed 250K and 6.0 SNP-A analyses on 140 patients with primary (p) and secondary (s) AML and correlated the results with clinical outcomes and Flt-3/nucleophosmin (NPM-1) status.
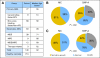
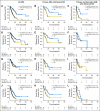
Results: SNP-A is more sensitive than MC in detecting unbalanced lesions (pAML, 65% v 39%, P = .002; and sAML, 78% v 51%, P = .003). Acquired somatic UPD, not detectable by MC, was common in our AML cohort (29% in pAML and 35% in sAML). Patients with SNP-A lesions including acquired somatic UPD exhibited worse overall survival (OS) and event-free survival (EFS) in pAML with normal MC and in pAML/sAML with abnormal MC. SNP-A improved the predictive value of Flt-3 internal tandem duplication/NPM-1 status, with inferior survival seen in patients with additional SNP-A defects. Multivariate analyses confirmed the independent predictive value of SNP-A defects for OS (hazard ratio [HR] = 2.52; 95% CI, 1.29 to 5.22; P = .006) and EFS (HR = 1.72; 95% CI, 1.12 to 3.48; P = .04).
Conclusion: SNP-A analysis allows enhanced detection of chromosomal abnormalities and provides important prognostic impact in AML.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures

References
-
- Schiffer CA, Lee EJ, Tomiyasu T, et al. Prognostic impact of cytogenetic abnormalities in patients with de novo acute nonlymphocytic leukemia. Blood. 1989;73:263–270. - PubMed
-
- Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. - PubMed
-
- Grimwade D, Walker H, Harrison G, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): Analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–1320. - PubMed
-
- Byrd JC, Mrozek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. - PubMed
-
- Smith SM, Le Beau MM, Huo D, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: The University of Chicago series. Blood. 2003;102:43–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

