Modeling discrete heterogeneity in X-ray diffraction data by fitting multi-conformers
- PMID: 19770508
- PMCID: PMC2756169
- DOI: 10.1107/S0907444909030613
Modeling discrete heterogeneity in X-ray diffraction data by fitting multi-conformers
Abstract
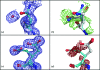
The native state of a protein is regarded to be an ensemble of conformers, which allows association with binding partners. While some of this structural heterogeneity is retained upon crystallization, reliably extracting heterogeneous features from diffraction data has remained a challenge. In this study, a new algorithm for the automatic modelling of discrete heterogeneity is presented. At high resolution, the authors' single multi-conformer model, with correlated structural features to represent heterogeneity, shows improved agreement with the diffraction data compared with a single-conformer model. The model appears to be representative of the set of structures present in the crystal. In contrast, below 2 A resolution representing ambiguous electron density by correlated multi-conformers in a single model does not yield better agreement with the experimental data. Consistent with previous studies, this suggests that variability in multi-conformer models at lower resolution levels reflects uncertainty more than coordinated motion.
Figures



References
-
- Afonine, P. V., Grosse-Kunstleve, R. W. & Adams, P. D. (2005b). CCP4 Newsl.42, contribution 8.
-
- Bedem, H. van den, Lotan, I., Latombe, J.-C. & Deacon, A. M. (2005). Acta Cryst. D61, 2–13. - PubMed
-
- Berman, H. M., Henrick, K., Nakamura, H. & Arnold, E. (2006). Nature Struct. Mol. Biol.13, 185.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

