Genetically encoded biosensors based on engineered fluorescent proteins
- PMID: 19771330
- PMCID: PMC3000468
- DOI: 10.1039/b907749a
Genetically encoded biosensors based on engineered fluorescent proteins
Abstract
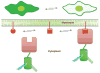
Fluorescent proteins have revolutionized cell biology by allowing researchers to non-invasively peer into the inner workings of cells and organisms. While the most common applications of fluorescent proteins are to image expression, localization, and dynamics of protein chimeras, there is a growing interest in using fluorescent proteins to create biosensors for minimally invasive imaging of concentrations of ions and small molecules, the activity of enzymes, and changes in the conformation of proteins in living cells. This tutorial review provides an overview of the progress made in the development of fluorescent protein-based biosensors to date.
Figures
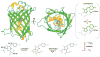
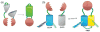
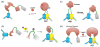

Similar articles
-
Protein biosensors based on the principle of fluorescence resonance energy transfer for monitoring cellular dynamics.Biotechnol Lett. 2006 Dec;28(24):1971-82. doi: 10.1007/s10529-006-9193-5. Epub 2006 Oct 5. Biotechnol Lett. 2006. PMID: 17021660 Review.
-
Temporal Metabolite, Ion, and Enzyme Activity Profiling Using Fluorescence Microscopy and Genetically Encoded Biosensors.Methods Mol Biol. 2019;1978:343-353. doi: 10.1007/978-1-4939-9236-2_21. Methods Mol Biol. 2019. PMID: 31119673 Free PMC article.
-
Novel Fluorescence-Based Biosensors Incorporating Unnatural Amino Acids.Methods Enzymol. 2017;589:191-219. doi: 10.1016/bs.mie.2017.01.012. Epub 2017 Feb 21. Methods Enzymol. 2017. PMID: 28336064
-
Fluorescence-Activating and Absorption-Shifting Tags for Advanced Imaging and Biosensing.Acc Chem Res. 2022 Nov 1;55(21):3125-3135. doi: 10.1021/acs.accounts.2c00098. Epub 2022 Oct 21. Acc Chem Res. 2022. PMID: 36269101 Review.
-
Design and application of genetically encoded biosensors.Trends Biotechnol. 2011 Mar;29(3):144-52. doi: 10.1016/j.tibtech.2010.12.004. Epub 2011 Jan 19. Trends Biotechnol. 2011. PMID: 21251723 Free PMC article. Review.
Cited by
-
Perspectives for using genetically encoded fluorescent biosensors in plants.Front Plant Sci. 2013 Jul 12;4:234. doi: 10.3389/fpls.2013.00234. eCollection 2013. Front Plant Sci. 2013. PMID: 23874345 Free PMC article.
-
Rapid construction of metabolite biosensors using domain-insertion profiling.Nat Commun. 2016 Jul 29;7:12266. doi: 10.1038/ncomms12266. Nat Commun. 2016. PMID: 27470466 Free PMC article.
-
Towards single-cell real-time imaging of energy metabolism in the brain.Front Neuroenergetics. 2010 Jun 7;2:4. doi: 10.3389/fnene.2010.00004. eCollection 2010. Front Neuroenergetics. 2010. PMID: 20577639 Free PMC article. No abstract available.
-
Red fluorescent proteins engineered from green fluorescent proteins.Proc Natl Acad Sci U S A. 2023 Nov 7;120(45):e2307687120. doi: 10.1073/pnas.2307687120. Epub 2023 Oct 23. Proc Natl Acad Sci U S A. 2023. PMID: 37871160 Free PMC article.
-
Proteins on the move: insights gained from fluorescent protein technologies.Nat Rev Mol Cell Biol. 2011 Sep 23;12(10):656-68. doi: 10.1038/nrm3199. Nat Rev Mol Cell Biol. 2011. PMID: 21941275 Review.
References
-
- Shimomura O, Johnson FH, Saiga Y. J Cell Comp Physiol. 1962;59:223–239. - PubMed
-
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Gene. 1992;111:229–233. - PubMed
-
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802–805. - PubMed
-
- Tsien RY. Annu Rev Biochem. 1998;67:509–544. - PubMed
-
- Miyawaki A, Llopis J, Heim R, McCaffery JM, et al. Nature. 1997;388:882–887. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

