Mechanisms of double-strand break repair in somatic mammalian cells
- PMID: 19772495
- PMCID: PMC2983087
- DOI: 10.1042/BJ20090942
Mechanisms of double-strand break repair in somatic mammalian cells
Erratum in
- Biochem J. 2010 Mar 15;426(3):389
Abstract
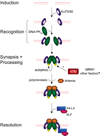
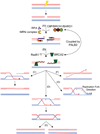
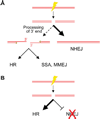
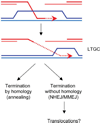
DNA chromosomal DSBs (double-strand breaks) are potentially hazardous DNA lesions, and their accurate repair is essential for the successful maintenance and propagation of genetic information. Two major pathways have evolved to repair DSBs: HR (homologous recombination) and NHEJ (non-homologous end-joining). Depending on the context in which the break is encountered, HR and NHEJ may either compete or co-operate to fix DSBs in eukaryotic cells. Defects in either pathway are strongly associated with human disease, including immunodeficiency and cancer predisposition. Here we review the current knowledge of how NHEJ and HR are controlled in somatic mammalian cells, and discuss the role of the chromatin context in regulating each pathway. We also review evidence for both co-operation and competition between the two pathways.
Figures
References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd Edition. Washington: ASM Press; 2006.
-
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. - PubMed
-
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. - PubMed
-
- Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

