Characterization of WRKY co-regulatory networks in rice and Arabidopsis
- PMID: 19772648
- PMCID: PMC2761919
- DOI: 10.1186/1471-2229-9-120
Characterization of WRKY co-regulatory networks in rice and Arabidopsis
Abstract
Background: The WRKY transcription factor gene family has a very ancient origin and has undergone extensive duplications in the plant kingdom. Several studies have pointed out their involvement in a range of biological processes, revealing that a large number of WRKY genes are transcriptionally regulated under conditions of biotic and/or abiotic stress. To investigate the existence of WRKY co-regulatory networks in plants, a whole gene family WRKYs expression study was carried out in rice (Oryza sativa). This analysis was extended to Arabidopsis thaliana taking advantage of an extensive repository of gene expression data.
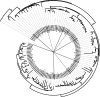
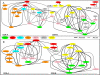
Results: The presented results suggested that 24 members of the rice WRKY gene family (22% of the total) were differentially-regulated in response to at least one of the stress conditions tested. We defined the existence of nine OsWRKY gene clusters comprising both phylogenetically related and unrelated genes that were significantly co-expressed, suggesting that specific sets of WRKY genes might act in co-regulatory networks. This hypothesis was tested by Pearson Correlation Coefficient analysis of the Arabidopsis WRKY gene family in a large set of Affymetrix microarray experiments. AtWRKYs were found to belong to two main co-regulatory networks (COR-A, COR-B) and two smaller ones (COR-C and COR-D), all including genes belonging to distinct phylogenetic groups. The COR-A network contained several AtWRKY genes known to be involved mostly in response to pathogens, whose physical and/or genetic interaction was experimentally proven. We also showed that specific co-regulatory networks were conserved between the two model species by identifying Arabidopsis orthologs of the co-expressed OsWRKY genes.
Conclusion: In this work we identified sets of co-expressed WRKY genes in both rice and Arabidopsis that are functionally likely to cooperate in the same signal transduction pathways. We propose that, making use of data from co-regulatory networks, it is possible to highlight novel clusters of plant genes contributing to the same biological processes or signal transduction pathways. Our approach will contribute to unveil gene cooperation pathways not yet identified by classical genetic analyses. This information will open new routes contributing to the dissection of WRKY signal transduction pathways in plants.
Figures


References
-
- Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol. 2004;55:853–867. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

