Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches
- PMID: 19773805
- PMCID: PMC2958924
- DOI: 10.1038/nrm2777
Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches
Abstract
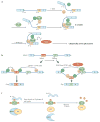
Alternative splicing of mRNA precursors provides an important means of genetic control and is a crucial step in the expression of most genes. Alternative splicing markedly affects human development, and its misregulation underlies many human diseases. Although the mechanisms of alternative splicing have been studied extensively, until the past few years we had not begun to realize fully the diversity and complexity of alternative splicing regulation by an intricate protein-RNA network. Great progress has been made by studying individual transcripts and through genome-wide approaches, which together provide a better picture of the mechanistic regulation of alternative pre-mRNA splicing.
Figures


References
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. - PubMed
-
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
-
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. - PubMed
-
- Nelson KK, Green MR. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989;3:1562–1571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

