Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins
- PMID: 19775250
- PMCID: PMC2784877
- DOI: 10.1111/j.1365-2958.2009.06891.x
Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins
Abstract
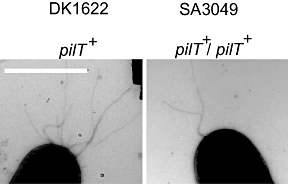
Type IV pili (T4P) are surface structures that undergo extension/retraction oscillations to generate cell motility. In Myxococcus xanthus, T4P are unipolarly localized and undergo pole-to-pole oscillations synchronously with cellular reversals. We investigated the mechanisms underlying these oscillations. We show that several T4P proteins localize symmetrically in clusters at both cell poles between reversals, and these clusters remain stationary during reversals. Conversely, the PilB and PilT motor ATPases that energize extension and retraction, respectively, localize to opposite poles with PilB predominantly at the piliated and PilT predominantly at the non-piliated pole, and these proteins oscillate between the poles during reversals. Therefore, T4P pole-to-pole oscillations involve the disassembly of T4P machinery at one pole and reassembly of this machinery at the opposite pole. Fluorescence recovery after photobleaching experiments showed rapid turnover of YFP-PilT in the polar clusters between reversals. Moreover, PilT displays bursts of accumulation at the piliated pole between reversals. These observations suggest that the spatial separation of PilB and PilT in combination with the noisy PilT accumulation at the piliated pole allow the temporal separation of extension and retraction. This is the first demonstration that the function of a molecular machine depends on disassembly and reassembly of its individual parts.
Figures







References
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;16:783–795. - PubMed
-
- Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol. 2006;61:1510–1522. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

