Ribosomal protein S12 and aminoglycoside antibiotics modulate A-site mRNA cleavage and transfer-messenger RNA activity in Escherichia coli
- PMID: 19776006
- PMCID: PMC2797289
- DOI: 10.1074/jbc.M109.062745
Ribosomal protein S12 and aminoglycoside antibiotics modulate A-site mRNA cleavage and transfer-messenger RNA activity in Escherichia coli
Abstract
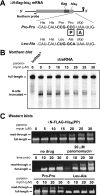
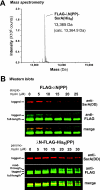
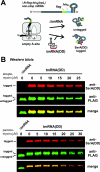
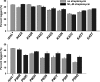
Translational pausing in Escherichia coli can lead to mRNA cleavage within the ribosomal A-site. A-site mRNA cleavage is thought to facilitate transfer-messenger RNA (tmRNA).SmpB- mediated recycling of stalled ribosome complexes. Here, we demonstrate that the aminoglycosides paromomycin and streptomycin inhibit A-site cleavage of stop codons during inefficient translation termination. Aminoglycosides also induced stop codon read-through, suggesting that these antibiotics alleviate ribosome pausing during termination. Streptomycin did not inhibit A-site cleavage in rpsL mutants, which express streptomycin-resistant variants of ribosomal protein S12. However, rpsL strains exhibited reduced A-site mRNA cleavage compared with rpsL(+) cells. Additionally, tmRNA.SmpB-mediated SsrA peptide tagging was significantly reduced in several rpsL strains but could be fully restored in a subset of mutants when treated with streptomycin. The streptomycin-dependent rpsL(P90K) mutant also showed significantly lower levels of A-site cleavage and tmRNA.SmpB activity. Mutations in rpsD (encoding ribosomal protein S4), which suppressed streptomycin dependence, were able to partially restore A-site cleavage to rpsL(P90K) cells but failed to increase tmRNA.SmpB activity. Taken together, these results show that perturbations to A-site structure and function modulate A-site mRNA cleavage and tmRNA.SmpB activity. We propose that tmRNA.SmpB binds to streptomycin-resistant rpsL ribosomes less efficiently, leading to a partial loss of ribosome rescue function in these mutants.
Figures





Similar articles
-
A-site mRNA cleavage is not required for tmRNA-mediated ssrA-peptide tagging.PLoS One. 2013 Nov 19;8(11):e81319. doi: 10.1371/journal.pone.0081319. eCollection 2013. PLoS One. 2013. PMID: 24260569 Free PMC article.
-
Kinetics of paused ribosome recycling in Escherichia coli.J Mol Biol. 2009 Nov 27;394(2):251-67. doi: 10.1016/j.jmb.2009.09.020. Epub 2009 Sep 15. J Mol Biol. 2009. PMID: 19761774 Free PMC article.
-
Various effects of paromomycin on tmRNA-directed trans-translation.J Biol Chem. 2003 Jul 25;278(30):27672-80. doi: 10.1074/jbc.M211724200. Epub 2003 May 15. J Biol Chem. 2003. PMID: 12754221
-
The tmRNA system for translational surveillance and ribosome rescue.Annu Rev Biochem. 2007;76:101-24. doi: 10.1146/annurev.biochem.75.103004.142733. Annu Rev Biochem. 2007. PMID: 17291191 Review.
-
A salvage pathway for protein structures: tmRNA and trans-translation.Annu Rev Microbiol. 2003;57:101-23. doi: 10.1146/annurev.micro.57.030502.090945. Epub 2003 May 1. Annu Rev Microbiol. 2003. PMID: 12730326 Review.
Cited by
-
The role of SmpB and the ribosomal decoding center in licensing tmRNA entry into stalled ribosomes.RNA. 2011 Sep;17(9):1727-36. doi: 10.1261/rna.2821711. Epub 2011 Jul 27. RNA. 2011. PMID: 21795410 Free PMC article.
-
A novel family of toxin/antitoxin proteins in Bacillus species.FEBS Lett. 2012 Jan 20;586(2):132-6. doi: 10.1016/j.febslet.2011.12.020. Epub 2011 Dec 22. FEBS Lett. 2012. PMID: 22200572 Free PMC article.
-
Restrictive Streptomycin Resistance Mutations Decrease the Formation of Attaching and Effacing Lesions in Escherichia coli O157:H7 Strains.Antimicrob Agents Chemother. 2013 Sep;57(9):4260-4266. doi: 10.1128/AAC.00709-13. Epub 2013 Jun 24. Antimicrob Agents Chemother. 2013. PMID: 23796920 Free PMC article.
-
Streptomycin Induced Stress Response in Salmonella enterica Serovar Typhimurium Shows Distinct Colony Scatter Signature.PLoS One. 2015 Aug 7;10(8):e0135035. doi: 10.1371/journal.pone.0135035. eCollection 2015. PLoS One. 2015. PMID: 26252374 Free PMC article.
-
Evolutionary trajectory of bacterial resistance to antibiotics and antimicrobial peptides in Escherichia coli.mSystems. 2025 Mar 18;10(3):e0170024. doi: 10.1128/msystems.01700-24. Epub 2025 Feb 27. mSystems. 2025. PMID: 40013796 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

