Adenosine deamination in human transcripts generates novel microRNA binding sites
- PMID: 19776031
- PMCID: PMC2778373
- DOI: 10.1093/hmg/ddp443
Adenosine deamination in human transcripts generates novel microRNA binding sites
Abstract
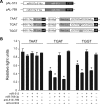
Animals regulate gene expression at multiple levels, contributing to the complexity of the proteome. Among these regulatory events are post-transcriptional gene silencing, mediated by small non-coding RNAs (e.g. microRNAs), and adenosine-to-inosine (A-to-I) editing, generated by adenosine deaminases that act on double-stranded RNA (ADAR). Recent data suggest that these regulatory processes are connected at a fundamental level. A-to-I editing can affect Drosha processing or directly alter the microRNA (miRNA) sequences responsible for mRNA targeting. Here, we analyzed the previously reported adenosine deaminations occurring in human cDNAs, and asked if there was a relationship between A-to-I editing events in the mRNA 3' untranslated regions (UTRs) and mRNA:miRNA binding. We find significant correlations between A-to-I editing and changes in miRNA complementarities. In all, over 3000 of the 12 723 distinct adenosine deaminations assessed were found to form 7-mer complementarities (known as seed matches) to a subset of human miRNAs. In 200 of the ESTs, we also noted editing within a specific 13 nucleotide motif. Strikingly, deamination of this motif simultaneously creates seed matches to three (otherwise unrelated) miRNAs. Our results suggest the creation of miRNA regulatory sites as a novel function for ADAR activity. Consequently, many miRNA target sites may only be identifiable through examining expressed sequences.
Figures




References
-
- Yoshida M., Kaziro Y., Ukita T. The modification of nucleosides and nucleotides. X. Evidence for the important role of inosine residue in codon recognition of yeast alanine tRNA. Biochim. Biophys. Acta. 1968;166:646–655. - PubMed
-
- Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., Sanders-Bush E., Emeson R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. - PubMed
-
- Li J.B., Levanon E.Y., Yoon J.K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., Church G.M. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. - PubMed
-
- Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

