miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression
- PMID: 19777054
- PMCID: PMC2743997
- DOI: 10.1371/journal.pone.0007158
miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression
Abstract
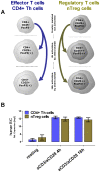
Background: In humans and mice naturally occurring CD4(+)CD25(+) regulatory T cells (nTregs) are a thymus-derived subset of T cells, crucial for the maintenance of peripheral tolerance by controlling not only potentially autoreactive T cells but virtually all cells of the adaptive and innate immune system. Recent work using Dicer-deficient mice irrevocably demonstrated the importance of miRNAs for nTreg cell-mediated tolerance.
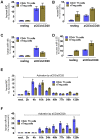
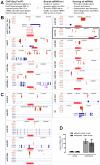
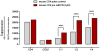
Principal findings: DNA-Microarray analyses of human as well as murine conventional CD4(+) Th cells and nTregs revealed a strong up-regulation of mature miR-155 (microRNA-155) upon activation in both populations. Studying miR-155 expression in FoxP3-deficient scurfy mice and performing FoxP3 ChIP-Seq experiments using activated human T lymphocytes, we show that the expression and maturation of miR-155 seem to be not necessarily regulated by FoxP3. In order to address the functional relevance of elevated miR-155 levels, we transfected miR-155 inhibitors or mature miR-155 RNAs into freshly-isolated human and mouse primary CD4(+) Th cells and nTregs and investigated the resulting phenotype in nTreg suppression assays. Whereas miR-155 inhibition in conventional CD4(+) Th cells strengthened nTreg cell-mediated suppression, overexpression of mature miR-155 rendered these cells unresponsive to nTreg cell-mediated suppression.
Conclusion: Investigation of FoxP3 downstream targets, certainly of bound and regulated miRNAs revealed the associated function between the master regulator FoxP3 and miRNAs as regulators itself. miR-155 is shown to be crucially involved in nTreg cell mediated tolerance by regulating the susceptibility of conventional human as well as murine CD4(+) Th cells to nTreg cell-mediated suppression.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

