Aberrant DNA methylation occurs in colon neoplasms arising in the azoxymethane colon cancer model
- PMID: 19777566
- PMCID: PMC2875385
- DOI: 10.1002/mc.20581
Aberrant DNA methylation occurs in colon neoplasms arising in the azoxymethane colon cancer model
Abstract
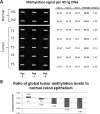
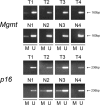
Mouse models of intestinal tumors have advanced our understanding of the role of gene mutations in colorectal malignancy. However, the utility of these systems for studying the role of epigenetic alterations in intestinal neoplasms remains to be defined. Consequently, we assessed the role of aberrant DNA methylation in the azoxymethane (AOM) rodent model of colon cancer. AOM induced tumors display global DNA hypomethylation, which is similar to human colorectal cancer. We next assessed the methylation status of a panel of candidate genes previously shown to be aberrantly methylated in human cancer or in mouse models of malignant neoplasms. This analysis revealed different patterns of DNA methylation that were gene specific. Zik1 and Gja9 demonstrated cancer-specific aberrant DNA methylation, whereas, Cdkn2a/p16, Igfbp3, Mgmt, Id4, and Cxcr4 were methylated in both the AOM tumors and normal colon mucosa. No aberrant methylation of Dapk1 or Mlt1 was detected in the neoplasms, but normal colon mucosa samples displayed methylation of these genes. Finally, p19(Arf), Tslc1, Hltf, and Mlh1 were unmethylated in both the AOM tumors and normal colon mucosa. Thus, aberrant DNA methylation does occur in AOM tumors, although the frequency of aberrantly methylated genes appears to be less common than in human colorectal cancer. Additional studies are necessary to further characterize the patterns of aberrantly methylated genes in AOM tumors.
2009 Wiley-Liss, Inc.
Figures


References
-
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. - PubMed
-
- Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annual Reviews Genomics and Human Genetics. 2002;3:101–128. - PubMed
-
- Janssen KP, Abala M, El Marjou F, Louvard D, Robine S. Mouse models of K-ras-initiated carcinogenesis. Biochim Biophys Acta. 2005;1756(2):145–154. - PubMed
-
- Munoz NM, Upton M, Rojas A, et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66(20):9837–9844. - PubMed
-
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11(11):1068–1075. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

