Critical role of glycosylation in determining the length and structure of T cell epitopes
- PMID: 19778434
- PMCID: PMC2760507
- DOI: 10.1186/1745-7580-5-4
Critical role of glycosylation in determining the length and structure of T cell epitopes
Abstract
Background: Using a combined in silico approach, we investigated the glycosylation of T cell epitopes and autoantigens. The present systems biology analysis was made possible by currently available databases (representing full proteomes, known human T cell epitopes and autoantigens) as well as glycosylation prediction tools.
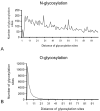
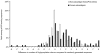
Results: We analyzed the probable glycosylation of human T cell epitope sequences extracted from the ImmuneEpitope Database. Our analysis suggests that in contrast to full length SwissProt entries, only a minimal portion of experimentally verified T cell epitopes is potentially N- or O-glycosylated (2.26% and 1.22%, respectively). Bayesian analysis of entries extracted from the Autoantigen Database suggests a correlation between N-glycosylation and autoantigenicity. The analysis of random generated sequences shows that glycosylation probability is also affected by peptide length. Our data suggest that the lack of peptide glycosylation, a feature that probably favors effective recognition by T cells, might have resulted in a selective advantage for short peptides to become T cell epitopes. The length of T cell epitopes is at the intersection of curves determining specificity and glycosylation probability. Thus, the range of length of naturally occurring T cell epitopes may ensure the maximum specificity with the minimal glycosylation probability.
Conclusion: The findings of this bioinformatical approach shed light on fundamental factors that might have shaped adaptive immunity during evolution. Our data suggest that amino acid sequence-based hypo/non-glycosylation of certain segments of proteins might be substantial for determining T cell immunity/autoimmunity.
Figures




References
-
- Ishioka GY, Lamont AG, Thomson D, Bulbow N, Gaeta FC, Sette A, Grey HM. MHC interaction and T cell recognition of carbohydrates and glycopeptides. J Immunol. 1992;148:2446–51. - PubMed
-
- Hanisch FG, Schwientek T, Von Bergwelt-Baildon MS, Schultze JL, Finn O. O-Linked glycans control glycoprotein processing by antigen-presenting cells: a biochemical approach to the molecular aspects of MUC1. processing by dendritic cells. Eur J Immunol. 2003;33:3242–54. doi: 10.1002/eji.200324189. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
