The rapamycin-regulated gene expression signature determines prognosis for breast cancer
- PMID: 19778445
- PMCID: PMC2761377
- DOI: 10.1186/1476-4598-8-75
The rapamycin-regulated gene expression signature determines prognosis for breast cancer
Abstract
Background: Mammalian target of rapamycin (mTOR) is a serine/threonine kinase involved in multiple intracellular signaling pathways promoting tumor growth. mTOR is aberrantly activated in a significant portion of breast cancers and is a promising target for treatment. Rapamycin and its analogues are in clinical trials for breast cancer treatment. Patterns of gene expression (metagenes) may also be used to simulate a biologic process or effects of a drug treatment. In this study, we tested the hypothesis that the gene-expression signature regulated by rapamycin could predict disease outcome for patients with breast cancer.
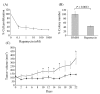
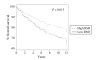
Results: Colony formation and sulforhodamine B (IC50 < 1 nM) assays, and xenograft animals showed that MDA-MB-468 cells were sensitive to treatment with rapamycin. The comparison of in vitro and in vivo gene expression data identified a signature, termed rapamycin metagene index (RMI), of 31 genes upregulated by rapamycin treatment in vitro as well as in vivo (false discovery rate of 10%). In the Miller dataset, RMI did not correlate with tumor size or lymph node status. High (>75th percentile) RMI was significantly associated with longer survival (P = 0.015). On multivariate analysis, RMI (P = 0.029), tumor size (P = 0.015) and lymph node status (P = 0.001) were prognostic. In van 't Veer study, RMI was not associated with the time to develop distant metastasis (P = 0.41). In the Wang dataset, RMI predicted time to disease relapse (P = 0.009).
Conclusion: Rapamycin-regulated gene expression signature predicts clinical outcome in breast cancer. This supports the central role of mTOR signaling in breast cancer biology and provides further impetus to pursue mTOR-targeted therapies for breast cancer treatment.
Figures


References
-
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. - PubMed
-
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

