Chemical probes identify a role for histone deacetylase 3 in Friedreich's ataxia gene silencing
- PMID: 19778726
- PMCID: PMC2909763
- DOI: 10.1016/j.chembiol.2009.07.010
Chemical probes identify a role for histone deacetylase 3 in Friedreich's ataxia gene silencing
Abstract
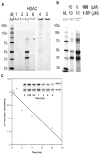
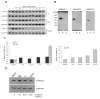
We recently identified a class of pimelic diphenylamide histone deacetylase (HDAC) inhibitors that show promise as therapeutics in the neurodegenerative diseases Friedreich's ataxia (FRDA) and Huntington's disease. Here, we describe chemical approaches to identify the HDAC enzyme target of these inhibitors. Incubation of a trifunctional activity-based probe with a panel of class I and class II recombinant HDAC enzymes, followed by click chemistry addition of a fluorescent dye and gel electrophoresis, identifies HDAC3 as a unique high-affinity target of the probe. Photoaffinity labeling in a nuclear extract prepared from human lymphoblasts with the trifunctional probe, followed by biotin addition through click chemistry, streptavidin enrichment, and Western blotting also identifies HDAC3 as the preferred cellular target of the inhibitor. Additional inhibitors with different HDAC specificity profiles were synthesized, and results from transcription experiments in FRDA cells point to a unique role for HDAC3 in gene silencing in Friedreich's ataxia.
Conflict of interest statement
J.M.G. is a consultant to Repligen Corporation, and has a competing financial interest in this work. H.P. and J.R.R. are employees of Repligen Corporation.
Figures




References
-
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1-11 in the rat brain. J Mol Neurosci. 2007;31:47–58. - PubMed
-
- Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharm Des. 2008;14:505–528. - PubMed
-
- Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem Rev. 2006;106:3279–3301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

