Two Dot1 isoforms in Saccharomyces cerevisiae as a result of leaky scanning by the ribosome
- PMID: 19778927
- PMCID: PMC2790890
- DOI: 10.1093/nar/gkp765
Two Dot1 isoforms in Saccharomyces cerevisiae as a result of leaky scanning by the ribosome
Abstract
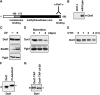
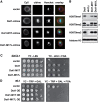
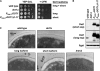
Dot1 is a conserved histone methyltransferase that methylates histone H3 on lysine 79. We previously observed that in Saccharomyces cerevisiae, a single DOT1 gene encodes two Dot1 protein species. Here, we show that the relative abundance of the two isoforms changed under nutrient-limiting conditions. A mutagenesis approach showed that the two Dot1 isoforms are produced from two alternative translation start sites as a result of leaky scanning by the ribosome. The leaky scanning was not affected by the 5'- or 3'-untranslated regions of DOT1, indicating that translation initiation is determined by the DOT1 coding sequence. Construction of yeast strains expressing either one of the isoforms showed that both were sufficient for Dot1's role in global H3K79 methylation and telomeric gene silencing. However, the absence of the long isoform of Dot1 altered the resistance of yeast cells to the chitin-binding drug Calcofluor White, suggesting that the two Dot1 isoforms have a differential function in cell wall biogenesis.
Figures


References
-
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. - PubMed
-
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. - PubMed
-
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

