Bayesian phylogeography finds its roots
- PMID: 19779555
- PMCID: PMC2740835
- DOI: 10.1371/journal.pcbi.1000520
Bayesian phylogeography finds its roots
Abstract
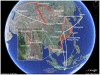
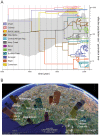
As a key factor in endemic and epidemic dynamics, the geographical distribution of viruses has been frequently interpreted in the light of their genetic histories. Unfortunately, inference of historical dispersal or migration patterns of viruses has mainly been restricted to model-free heuristic approaches that provide little insight into the temporal setting of the spatial dynamics. The introduction of probabilistic models of evolution, however, offers unique opportunities to engage in this statistical endeavor. Here we introduce a Bayesian framework for inference, visualization and hypothesis testing of phylogeographic history. By implementing character mapping in a Bayesian software that samples time-scaled phylogenies, we enable the reconstruction of timed viral dispersal patterns while accommodating phylogenetic uncertainty. Standard Markov model inference is extended with a stochastic search variable selection procedure that identifies the parsimonious descriptions of the diffusion process. In addition, we propose priors that can incorporate geographical sampling distributions or characterize alternative hypotheses about the spatial dynamics. To visualize the spatial and temporal information, we summarize inferences using virtual globe software. We describe how Bayesian phylogeography compares with previous parsimony analysis in the investigation of the influenza A H5N1 origin and H5N1 epidemiological linkage among sampling localities. Analysis of rabies in West African dog populations reveals how virus diffusion may enable endemic maintenance through continuous epidemic cycles. From these analyses, we conclude that our phylogeographic framework will make an important asset in molecular epidemiology that can be easily generalized to infer biogeogeography from genetic data for many organisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
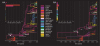

 , for the gamma priors on the rates; either
, for the gamma priors on the rates; either  , where
, where  is an arbitrary constant, or
is an arbitrary constant, or  , where
, where  is the distance between location
is the distance between location  and
and  . A truncated Poisson (TP) prior with
. A truncated Poisson (TP) prior with  was used in the Bayesian stochastic search variable selection (BSSVS) procedure.
was used in the Bayesian stochastic search variable selection (BSSVS) procedure.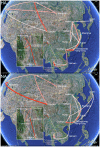

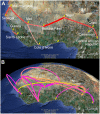

References
-
- Holmes E. The phylogeography of human viruses. Molecular Ecology. 2004;13:745–756. - PubMed
-
- Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. - PubMed
-
- Olsen SJ, Chang HL, Cheung TYY, Tang AFY, Fisk TL, et al. Transmission of the severe acute respiratory syndrome on aircraft. N Engl J Med. 2003;349:2416–2422. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

