Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL)
- PMID: 19779621
- PMCID: PMC2745703
- DOI: 10.1371/journal.pone.0007169
Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL)
Abstract
Background: MicroRNAs (miRNAs) are short, noncoding RNAs that regulate the expression of multiple target genes. Deregulation of miRNAs is common in human tumorigenesis. The miRNAs, MIR-15a/16-1, at chromosome band 13q14 are down-regulated in the majority of patients with chronic lymphocytic leukaemia (CLL).
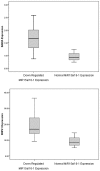
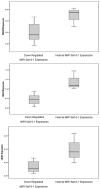
Methodology/principal findings: We have measured the expression of MIR-15a/16-1, and 92 computationally-predicted MIR-15a/16-1 target genes in CLL patients and in normal controls. We identified 35 genes that are deregulated in CLL patients, 5 of which appear to be specific targets of the MIR-15a/16-1 cluster. These targets included 2 genes (BAZ2A and RNF41) that were significantly up-regulated (p<0.05) and 3 genes (RASSF5, MKK3 and LRIG1) that were significantly down-regulated (p<0.05) in CLL patients with down-regulated MIR-15a/16-1 expression.
Significance: The genes identified here as being subject to MIR-15a/16-1 regulation could represent direct or indirect targets of these miRNAs. Many of these are good biological candidates for involvement in tumorigenesis and as such, may be important in the aetiology of CLL.
Conflict of interest statement
Figures

Similar articles
-
Expression of miR-15a and miR-16-1 in patients with chronic lymphocytic leukemia.Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2013 Dec;157(4):284-93. doi: 10.5507/bp.2013.057. Epub 2013 Sep 5. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2013. PMID: 24026141
-
Aberrant microRNA expression in Chinese patients with chronic lymphocytic leukemia.Leuk Res. 2011 Jun;35(6):730-4. doi: 10.1016/j.leukres.2010.11.005. Epub 2010 Dec 3. Leuk Res. 2011. PMID: 21130495
-
Therapeutic implications of activation of the host gene (Dleu2) promoter for miR-15a/16-1 in chronic lymphocytic leukemia.Oncogene. 2014 Jun 19;33(25):3307-15. doi: 10.1038/onc.2013.291. Epub 2013 Sep 2. Oncogene. 2014. PMID: 23995789 Free PMC article.
-
Role of miR-15/16 in CLL.Cell Death Differ. 2015 Jan;22(1):6-11. doi: 10.1038/cdd.2014.87. Epub 2014 Jun 27. Cell Death Differ. 2015. PMID: 24971479 Free PMC article. Review.
-
Evaluation of MiR-15a and MiR-16-1 as prognostic biomarkers in chronic lymphocytic leukemia.Biomed Pharmacother. 2017 Aug;92:864-869. doi: 10.1016/j.biopha.2017.05.144. Epub 2017 Jun 6. Biomed Pharmacother. 2017. PMID: 28599250 Review.
Cited by
-
Non random distribution of genomic features in breakpoint regions involved in chronic myeloid leukemia cases with variant t(9;22) or additional chromosomal rearrangements.Mol Cancer. 2010 May 25;9:120. doi: 10.1186/1476-4598-9-120. Mol Cancer. 2010. PMID: 20500819 Free PMC article.
-
The Role of MicroRNAs in Kidney Disease.Noncoding RNA. 2015 Nov 18;1(3):192-221. doi: 10.3390/ncrna1030192. Noncoding RNA. 2015. PMID: 29861424 Free PMC article. Review.
-
Intracellular and Extracellular miRNAs in Regulation of Angiogenesis Signaling.Curr Angiogenes. 2012 Dec;4(102):299-307. doi: 10.2174/2211552811201040299. Curr Angiogenes. 2012. PMID: 23914347 Free PMC article.
-
MicroRNA-16 inhibits migration and invasion via regulation of the Wnt/β-catenin signaling pathway in ovarian cancer.Oncol Lett. 2019 Mar;17(3):2631-2638. doi: 10.3892/ol.2019.9923. Epub 2019 Jan 14. Oncol Lett. 2019. PMID: 30854038 Free PMC article.
-
Interpreting the MicroRNA-15/107 family: interaction identification by combining network based and experiment supported approach.BMC Med Genet. 2019 May 31;20(1):96. doi: 10.1186/s12881-019-0824-9. BMC Med Genet. 2019. PMID: 31151434 Free PMC article.
References
-
- Rozman C, Montserrat E. Chronic lymphocytic leukemia. N Engl J Med. 1995;333:1052–1057. - PubMed
-
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. - PubMed
-
- Facon T, Avet-Loiseau H, Guillerm G, Moreau P, Genevieve F, et al. Chromosome 13 abnormalities identified by FISH analysis and serum beta2-microglobulin produce a powerful myeloma staging system for patients receiving high-dose therapy. Blood. 2001;97:1566–1571. - PubMed
-
- La Starza R, Wlodarska I, Aventin A, Falzetti D, Crescenzi B, et al. Molecular delineation of 13q deletion boundaries in 20 patients with myeloid malignancies. Blood. 1998;91:231–237. - PubMed
-
- Schlade-Bartusiak K, Stembalska A, Ramsey D. Significant involvement of chromosome 13q deletions in progression of larynx cancer, detected by comparative genomic hybridization. J Appl Genet. 2005;46:407–413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

