Parallel changes in gene expression in peripheral blood mononuclear cells and the brain after maternal separation in the mouse
- PMID: 19781058
- PMCID: PMC2759952
- DOI: 10.1186/1756-0500-2-195
Parallel changes in gene expression in peripheral blood mononuclear cells and the brain after maternal separation in the mouse
Abstract
Background: The functional integration of the neuro-, endocrine- and immune-systems suggests that the transcriptome of white blood cells may reflect neuropsychiatric states, and be used as a non-invasive diagnostic indicator. We used a mouse maternal separation model, a paradigm of early adversity, to test the hypothesis that transcriptional changes in peripheral blood mononuclear cells (PBMCs) are paralleled by specific gene expression changes in prefrontal cortex (PFC), hippocampus (Hic) and hypothalamus (Hyp). Furthermore, we evaluated whether gene expression profiles of PBMCs could be used to predict the separation status of individual animals.
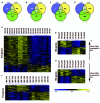
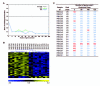
Findings: Microarray gene expression profiles of all three brain regions provided substantial evidence of stress-related neural differences between maternally separated and control animals. For example, changes in expression of genes involved in the glutamatergic and GABAergic systems were identified in the PFC and Hic, supporting a stress-related hyperglutamatergic state within the separated group. The expression of 50 genes selected from the PBMC microarray data provided sufficient information to predict treatment classes with 95% accuracy. Importantly, stress-related transcriptome differences in PBMC populations were paralleled by stress-related gene expression changes in CNS target tissues.
Conclusion: These results confirm that the transcriptional profiles of peripheral immune tissues occur in parallel to changes in the brain and contain sufficient information for the efficient diagnostic prediction of stress-related neural states in mice. Future studies will need to evaluate the relevance of the predictor set of 50 genes within clinical settings, specifically within a context of stress-related disorders.
Figures



References
-
- Tsuang MT, Nossova N, Yager T, Tsuang MM, Guo SC, Shyu KG, Glatt SJ, Liew CC. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:1–5. doi: 10.1002/ajmg.b.30161. - DOI - PubMed
-
- Besedovsky HO, del Rey A. Immune-neuro-endocrine interactions: facts and hypotheses. Endocr Rev. 1996;17:64–102. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

