Characterisation of a GII-4 norovirus variant-specific surface-exposed site involved in antibody binding
- PMID: 19781066
- PMCID: PMC2762976
- DOI: 10.1186/1743-422X-6-150
Characterisation of a GII-4 norovirus variant-specific surface-exposed site involved in antibody binding
Abstract
Background: The human noroviruses are a highly diverse group of viruses with a single-stranded RNA genome encoding a single major structural protein (VP1), which has a hypervariable domain (P2 domain) as the most exposed part of the virion. The noroviruses are classified on the basis of nucleotide sequence diversity in the VP1-encoding ORF2 gene, which divides the majority of human noroviruses into two genogroups (GI and GII). GII-4 noroviruses are the major aetiological agent of outbreaks of gastroenteritis around the world. During a winter season the diversity among the GII-4 noroviruses has been shown to fluctuate, driving the appearance of new virus variants in the population. We have previously shown that sequence data and in silico modelling experiments suggest there are two surface-exposed sites (site A and site B) in the hypervariable P2 domain. We predict these sites may form a functional variant-specific epitope that evolves under selective pressure from the host immune response and gives rise to antibody escape mutants.
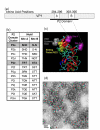
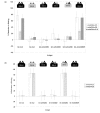
Results: In this paper, we describe the construction of recombinant baculoviruses to express VLPs representing one pre-epidemic and one epidemic variant of GII-4 noroviruses, and the production of monoclonal antibodies against them. We use these novel reagents to provide evidence that site A and site B form a conformational, variant-specific, surface-exposed site on the GII-4 norovirus capsid that is involved in antibody binding.
Conclusion: As predicted by our earlier study, significant amino acid changes at site A and site B give rise to GII-4 norovirus epidemic variants that are antibody escape mutants.
Figures

References
-
- Hay AJ, Zambon MC, Wolstenholme AJ, Skehel JJ, Smith MH. Molecular basis of resistance of influenza A viruses to amantadine. J Antimicrob Chemother. 1986;18:19–29. - PubMed
-
- Lambkin R, McLain L, Jones SE, Aldridge SL, Dimmock NJ. Neutralization escape mutants of type A influenza virus are readily selected by antisera from mice immunized with whole virus: a possible mechanism for antigenic drift. J Gen Virol. 1994;75:3493–3502. doi: 10.1099/0022-1317-75-12-3493. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

