Labeling strategies for 13C-detected aligned-sample solid-state NMR of proteins
- PMID: 19781966
- PMCID: PMC2804798
- DOI: 10.1016/j.jmr.2009.08.012
Labeling strategies for 13C-detected aligned-sample solid-state NMR of proteins
Abstract
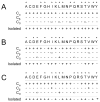
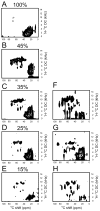
(13)C-detected solid-state NMR experiments have substantially higher sensitivity than the corresponding (15)N-detected experiments on stationary, aligned samples of isotopically labeled proteins. Several methods for tailoring the isotopic labeling are described that result in spatially isolated (13)C sites so that dipole-dipole couplings among the (13)C are minimized, thus eliminating the need for homonuclear (13)C-(13)C decoupling in either indirect or direct dimensions of one- or multi-dimensional NMR experiments that employ (13)C detection. The optimal percentage for random fractional (13)C labeling is between 25% and 35%. Specifically labeled glycerol and glucose can be used at the carbon sources to tailor the isotopic labeling, and the choice depends on the resonances of interest for a particular study. For investigations of the protein backbone, growth of the bacteria on [2-(13)C]-glucose-containing media was found to be most effective.
Figures

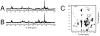






References
-
- Cross TA, DiVerdi JA, Opella SJ. Strategy for nitrogen NMR analysis of biopolymers. J Am Chem Soc. 1982;104:1759–61.
-
- Gu Z, Opella SJ. Three-dimensional 13C Shift/1H-15N Coupling/15N Shift solid-state NMR correlation spectroscopy. J Magn Reson. 138:193–198. - PubMed
-
- Gu Z, Opella SJ. Two- and three- dimensional 1H/13C PISEMA experiments and their application to backbone and side chain sites of amino acids and peptides. J Magn Reson. 1999b;140:340–346. - PubMed
-
- Schneider DM, Tycko R, Opella SJ. High-resolution solid-state triple nuclear magnetic resonance measurement of 13C-15N dipole-dipole couplings. J Magn Reson. 1987;73:568–573.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

