Transcription dynamics
- PMID: 19782025
- PMCID: PMC6326382
- DOI: 10.1016/j.molcel.2009.09.005
Transcription dynamics
Abstract
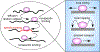
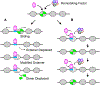
All aspects of transcription and its regulation involve dynamic events. The basal transcription machinery and regulatory components are dynamically recruited to their target genes, and dynamic interactions of transcription factors with chromatin--and with each other--play a key role in RNA polymerase assembly, initiation, and elongation. These short-term binding dynamics of transcription factors are superimposed by long-term cyclical behavior of chromatin opening and transcription factor-binding events. Its dynamic nature is not only a fundamental property of the transcription machinery, but it is emerging as an important modulator of physiological processes, particularly in differentiation and development.
Figures

References
-
- Araki R, Fukumura R, Sasaki N, Kasama Y, Suzuki N, Takahashi H, Tabata Y, Saito T, and Abe M (2006). More than 40,000 transcripts, including novel and noncoding transcripts, in mouse embryonic stem cells. Stem Cells 24, 2522–2528. - PubMed
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, and Fisher AG (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol 8, 532–538. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

