Reconstitution of heterochromatin-dependent transcriptional gene silencing
- PMID: 19782027
- PMCID: PMC2842978
- DOI: 10.1016/j.molcel.2009.07.030
Reconstitution of heterochromatin-dependent transcriptional gene silencing
Abstract
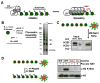
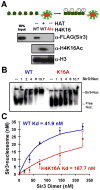
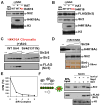
Heterochromatin assembly in budding yeast requires the SIR complex, which contains the NAD-dependent deacetylase Sir2 and the Sir3 and Sir4 proteins. Sir3 binds to nucleosomes containing deacetylated histone H4 lysine 16 (H4K16) and, with Sir4, promotes spreading of Sir2 and deacetylation along the chromatin fiber. Combined action of histone modifying and binding activities is a conserved hallmark of heterochromatin, but the relative contribution of each activity to silencing has remained unclear. Here, we reconstitute SIR-chromatin complexes using purified components and show that the SIR complex efficiently deacetylates chromatin templates and promotes the assembly of altered structures that silence Gal4-VP16-activated transcription. Silencing requires all three Sir proteins, even with fully deacetylated chromatin, and involves the specific association of Sir3 with deacetylated H4K16. These results define a minimal set of components that mediate heterochromatic gene silencing and demonstrate distinct contributions for histone deacetylation and nucleosome binding in the silencing mechanism.
Figures




References
-
- Chang JF, Hall BE, Tanny JC, Moazed D, Filman D, Ellenberger T. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure. 2003;11:637–649. - PubMed
-
- Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

