Systems biology asks new questions about sex differences
- PMID: 19783453
- PMCID: PMC2787703
- DOI: 10.1016/j.tem.2009.06.007
Systems biology asks new questions about sex differences
Abstract
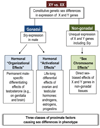
Females and males differ in physiology and in the incidence and progression of diseases. The sex-biased proximate factors causing sex differences in phenotype include direct effects of gonadal hormones and of genes represented unequally in the genome because of their X- or Y-linkage. Novel systems approaches have begun to assess the magnitude and character of sex differences in organization of gene networks on a genome-wide scale. These studies identify functionally related modules of genes that are coexpressed differently in males and females, and sites in the genome that regulate gene networks in a sex-specific manner. Measurement of the aggregate behavior of genes uncovers novel sex differences that can be related more effectively to susceptibility to disease.
Figures

References
-
- Perrot-Sinal TS. Do these genes make me look fat? Endocrinol. 2009;150:1075–1077. - PubMed
-
- Arnold AP. Sex chromosomes and brain gender. Nat. Rev. Neurosci. 2004;5:701–708. - PubMed
-
- Becker JB, et al. Strategies and methods for research on sex differences in brain and behavior. Endocrinol. 2005;146:1650–1673. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

