Comparative genomics of ethanolamine utilization
- PMID: 19783625
- PMCID: PMC2786565
- DOI: 10.1128/JB.00838-09
Comparative genomics of ethanolamine utilization
Abstract
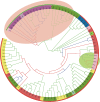
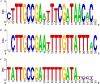
Ethanolamine can be used as a source of carbon and nitrogen by phylogenetically diverse bacteria. Ethanolamine-ammonia lyase, the enzyme that breaks ethanolamine into acetaldehyde and ammonia, is encoded by the gene tandem eutBC. Despite extensive studies of ethanolamine utilization in Salmonella enterica serovar Typhimurium, much remains to be learned about EutBC structure and catalytic mechanism, about the evolutionary origin of ethanolamine utilization, and about regulatory links between the metabolism of ethanolamine itself and the ethanolamine-ammonia lyase cofactor adenosylcobalamin. We used computational analysis of sequences, structures, genome contexts, and phylogenies of ethanolamine-ammonia lyases to address these questions and to evaluate recent data-mining studies that have suggested an association between bacterial food poisoning and the diol utilization pathways. We found that EutBC evolution included recruitment of a TIM barrel and a Rossmann fold domain and their fusion to N-terminal alpha-helical domains to give EutB and EutC, respectively. This fusion was followed by recruitment and occasional loss of auxiliary ethanolamine utilization genes in Firmicutes and by several horizontal transfers, most notably from the firmicute stem to the Enterobacteriaceae and from Alphaproteobacteria to Actinobacteria. We identified a conserved DNA motif that likely represents the EutR-binding site and is shared by the ethanolamine and cobalamin operons in several enterobacterial species, suggesting a mechanism for coupling the biosyntheses of apoenzyme and cofactor in these species. Finally, we found that the food poisoning phenotype is associated with the structural components of metabolosome more strongly than with ethanolamine utilization genes or with paralogous propanediol utilization genes per se.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

