DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit
- PMID: 19783643
- PMCID: PMC2785967
- DOI: 10.1104/pp.109.146985
DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit
Abstract
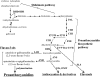
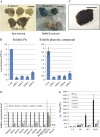
Proanthocyanidins (PAs) are secondary metabolites that contribute to the protection of the plant and also to the taste of the fruit, mainly through astringency. Persimmon (Diospyros kaki) is unique in being able to accumulate abundant PAs in the fruit flesh. Fruits of the nonastringent (NA)-type mutants lose their ability to produce PA at an early stage of fruit development, while those of the normal astringent (A) type remain rich in PA until fully ripened. The expression of many PA pathway genes was coincidentally terminated in the NA type at an early stage of fruit development. The five genes encoding the Myb transcription factor were isolated from an A-type cultivar (Kuramitsu). One of them, DkMyb4, showed an expression pattern synchronous to that of the PA pathway genes in A- and NA-type fruit flesh. The ectopic expression of DkMyb4 in kiwifruit (Actinidia deliciosa) induced PA biosynthesis but not anthocyanin biosynthesis. The suppression of DkMyb4 in persimmon calluses caused a substantial down-regulation of the PA pathway genes and PA biosynthesis. Furthermore, analysis of the DNA-binding ability of DkMyb4 showed that it directly binds to the MYBCORE cis-motif in the promoters of the some PA pathway genes. All our results indicate that DkMyb4 acts as a regulator of PA biosynthesis in persimmon and, therefore, suggest that the reduction in the DkMyb4 expression causes the NA-type-specific down-regulation of PA biosynthesis and resultant NA trait.
Figures







References
-
- Aharoni A, De Vos CHR, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O'Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28 319–332 - PubMed
-
- Akagi T, Ikegami A, Suzuki Y, Yoshida J, Yamada M, Sato A, Yonemori K (2009) Expression balances of structural genes in shikimate and flavonoid biosynthesis cause a difference in proanthocyanidin accumulation in persimmon (Diospyros kaki Thunb.) fruit. Planta 230 899–915 - PubMed
-
- Aron PM, Kennedy JA (2008) Flavan-3-ols: nature, occurrence and biological activity. Mol Nutr Food Res 52 79–104 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

