Untranslated regions of a mobile transcript mediate RNA metabolism
- PMID: 19783647
- PMCID: PMC2785979
- DOI: 10.1104/pp.109.144428
Untranslated regions of a mobile transcript mediate RNA metabolism
Abstract
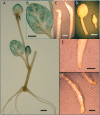
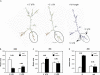
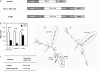
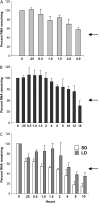
BEL1-like transcription factors are ubiquitous in plants and interact with KNOTTED1 types to regulate numerous developmental processes. In potato (Solanum tuberosum subsp. andigena), the BEL1-like transcription factor StBEL5 and its Knox protein partner regulate tuber formation by targeting genes that control growth. RNA detection methods and heterografting experiments demonstrated that StBEL5 transcripts are present in phloem cells and move across a graft union to localize in stolon tips, the site of tuber induction. This movement of RNA originates in leaf veins and petioles and is induced by a short-day photoperiod, regulated by the untranslated regions, and correlated with enhanced tuber production. Assays for RNA mobility suggest that both 5' and 3' untranslated regions contribute to the preferential accumulation of the StBEL5 RNA but that the 3' untranslated region may contribute more to transport from the leaf to the stem and into the stolons. Addition of the StBEL5 untranslated regions to another BEL1-like mRNA resulted in its preferential transport to stolon tips and enhanced tuber production. Transcript stability assays showed that the untranslated regions and a long-day photoperiod enhanced StBEL5 RNA stability in shoot tips. Upon fusion of the untranslated regions of StBEL5 to a beta-glucuronidase marker, translation in tobacco (Nicotiana tabacum) protoplasts was repressed by those constructs containing the 3' untranslated sequence. These results demonstrate that the untranslated regions of the mRNA of StBEL5 are involved in mediating its long-distance transport, in maintaining transcript stability, and in controlling translation.
Figures



References
-
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophot Int 11 36–42
-
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32 401–408 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

