Genetic evidence that the differential expression of the ligand-independent isoform of CTLA-4 is the molecular basis of the Idd5.1 type 1 diabetes region in nonobese diabetic mice
- PMID: 19783679
- PMCID: PMC2871291
- DOI: 10.4049/jimmunol.0802610
Genetic evidence that the differential expression of the ligand-independent isoform of CTLA-4 is the molecular basis of the Idd5.1 type 1 diabetes region in nonobese diabetic mice
Abstract
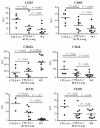
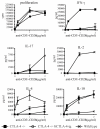
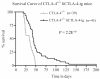
Idd5.1 regulates T1D susceptibility in nonobese diabetic (NOD) mice and has two notable candidate genes, Ctla4 and Icos. Reduced expression of one of the four CTLA-4 isoforms, ligand-independent CTLA-4 (liCTLA-4), which inhibits in vitro T cell activation and cytokine production similarly to full-length CTLA-4 (flCTLA-4), has been hypothesized to increase type 1 diabetes (T1D) susceptibility. However, further support of this hypothesis is required since the Idd5.1 haplotypes of the diabetes-susceptible NOD and the resistant B10 strains differ throughout Ctla4 and Icos. Using haplotype analysis and the generation of novel Idd5.1-congenic strains that differ at the disease-associated Ctla4 exon 2 single-nucleotide polymorphism, we demonstrate that increased expression of liCTLA-4 correlates with reduced T1D susceptibility. To directly assess the ability of liCTLA-4 to modulate T1D, we generated liCTLA-4-transgenic NOD mice and compared their diabetes susceptibility to nontransgenic littermates. NOD liCTLA-4-transgenic mice were protected from T1D to the same extent as NOD.B10 Idd5.1-congenic mice, demonstrating that increased liCTLA-4 expression alone can account for disease protection. To further investigate the in vivo function of liCTLA-4, specifically whether liCTLA-4 can functionally replace flCTLA-4 in vivo, we expressed the liCTLA-4 transgene in CTLA-4(-/-) B6 mice. CTLA-4(-/-) mice expressing liCTLA-4 accumulated fewer activated effector/memory CD4(+) T cells than CTLA-4(-/-) mice and the transgenic mice were partially rescued from the multiorgan inflammation and early lethality caused by the disruption of Ctla4. These results suggest that liCTLA-4 can partially replace some functions of flCTLA-4 in vivo and that this isoform evolved to reinforce the function of flCTLA-4.
Figures





References
-
- Leiter EH. Nonobese diabetic mice and the genetics of diabetes susceptibility. Curr Diab Rep. 2005;5:141–148. - PubMed
-
- Maier LM, Wicker LS. Genetic susceptibility to type 1 diabetes. Curr Opin Immunol. 2005;17:601–608. - PubMed
-
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. - PMC - PubMed
-
- Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B, Jones R, Ring SM, McArdle W, Pembrey ME, Strachan DP, Dunger DB, Twells RC, Clayton DG, Todd JA. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:773–779. - PMC - PubMed
-
- Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, Vella A, Nutland S, Rance HE, Maier L, Barratt BJ, Guja C, Ionescu-Tirgoviste C, Savage DA, Dunger DB, Widmer B, Strachan DP, Ring SM, Walker N, Clayton DG, Twells RC, Gough SC, Todd JA. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004;53:3020–3023. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

