The HSV-1 ICP27 RGG box specifically binds flexible, GC-rich sequences but not G-quartet structures
- PMID: 19783816
- PMCID: PMC2790906
- DOI: 10.1093/nar/gkp793
The HSV-1 ICP27 RGG box specifically binds flexible, GC-rich sequences but not G-quartet structures
Abstract
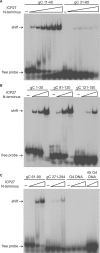
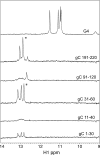
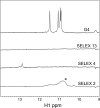
Herpes simplex virus 1 (HSV-1) protein ICP27, an important regulator for viral gene expression, directly recognizes and exports viral RNA through an N-terminal RGG box RNA binding motif, which is necessary and sufficient for RNA binding. An ICP27 N-terminal peptide, including the RGG box RNA binding motif, was expressed and its binding specificity was analyzed using EMSA and SELEX. DNA oligonucleotides corresponding to HSV-1 glycoprotein C (gC) mRNA, identified in a yeast three-hybrid analysis, were screened for binding to the ICP27 N-terminal peptide in EMSA experiments. The ICP27 N-terminus was able to bind most gC substrates. Notably, the ICP27 RGG box was unable to bind G-quartet structures recognized by the RGG domains of other proteins. SELEX analysis identified GC-rich RNA sequences as a common feature of recognition. NMR analysis of SELEX and gC sequences revealed that sequences able to bind to ICP27 did not form secondary structures and conversely, sequences that were not able to bind to ICP27 gave spectra consistent with base-pairing. Therefore, the ICP27 RGG box is unique in its recognition of nucleic acid sequences compared to other RGG box proteins; it prefers flexible, GC-rich substrates that do not form stable secondary structures.
Figures



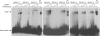

References
-
- Ingram A, Phelan A, Dunlop J, Clements JB. Immediate early protein IE63 of herpes simplex virus type 1 binds RNA directly. J. Gen. Virol. 1996;77((Pt 8)):1847–1851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

