A directed evolution design of a GCG-specific DNA hemimethylase
- PMID: 19783820
- PMCID: PMC2790894
- DOI: 10.1093/nar/gkp772
A directed evolution design of a GCG-specific DNA hemimethylase
Abstract
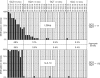
DNA cytosine-5 methyltransferases (C5-MTases) are valuable models to study sequence-specific modification of DNA and are becoming increasingly important tools for biotechnology. Here we describe a structure-guided rational protein design combined with random mutagenesis and selection to change the specificity of the HhaI C5-MTase from GCGC to GCG. The specificity change was brought about by a five-residue deletion and introduction of two arginine residues within and nearby one of the target recognizing loops. DNA protection assays, bisulfite sequencing and enzyme kinetics showed that the best selected variant is comparable to wild-type M.HhaI in terms of sequence fidelity and methylation efficiency, and supersedes the parent enzyme in transalkylation of DNA using synthetic cofactor analogs. The designed C5-MTase can be used to produce hemimethylated CpG sites in DNA, which are valuable substrates for studies of mammalian maintenance MTases.
Figures


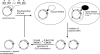


Similar articles
-
A 7-Deazaadenosylaziridine Cofactor for Sequence-Specific Labeling of DNA by the DNA Cytosine-C5 Methyltransferase M.HhaI.Molecules. 2015 Nov 23;20(11):20805-22. doi: 10.3390/molecules201119723. Molecules. 2015. PMID: 26610450 Free PMC article.
-
Mutational analysis of conserved residues in HhaI DNA methyltransferase.Nucleic Acids Res. 2002 Jun 15;30(12):2628-38. doi: 10.1093/nar/gkf380. Nucleic Acids Res. 2002. PMID: 12060679 Free PMC article.
-
Engineering the DNA cytosine-5 methyltransferase reaction for sequence-specific labeling of DNA.Nucleic Acids Res. 2012 Dec;40(22):11594-602. doi: 10.1093/nar/gks914. Epub 2012 Oct 5. Nucleic Acids Res. 2012. PMID: 23042683 Free PMC article.
-
Structure, function, and mechanism of HhaI DNA methyltransferases.Crit Rev Biochem Mol Biol. 2002;37(3):167-97. doi: 10.1080/10409230290771492. Crit Rev Biochem Mol Biol. 2002. PMID: 12139442 Review.
-
DNA methyltransferases: mechanistic models derived from kinetic analysis.Crit Rev Biochem Mol Biol. 2012 Mar-Apr;47(2):97-193. doi: 10.3109/10409238.2011.620942. Epub 2012 Jan 20. Crit Rev Biochem Mol Biol. 2012. PMID: 22260147 Review.
Cited by
-
Metadynamics simulation study on the conformational transformation of HhaI methyltransferase: an induced-fit base-flipping hypothesis.Biomed Res Int. 2014;2014:304563. doi: 10.1155/2014/304563. Epub 2014 Jun 19. Biomed Res Int. 2014. PMID: 25045662 Free PMC article.
-
Directed evolution of improved zinc finger methyltransferases.PLoS One. 2014 May 8;9(5):e96931. doi: 10.1371/journal.pone.0096931. eCollection 2014. PLoS One. 2014. PMID: 24810747 Free PMC article.
-
Conversion of the CG specific M.MpeI DNA methyltransferase into an enzyme predominantly methylating CCA and CCC sites.Nucleic Acids Res. 2024 Feb 28;52(4):1896-1908. doi: 10.1093/nar/gkad1217. Nucleic Acids Res. 2024. PMID: 38164970 Free PMC article.
-
DNA Labeling Using DNA Methyltransferases.Adv Exp Med Biol. 2016;945:511-535. doi: 10.1007/978-3-319-43624-1_19. Adv Exp Med Biol. 2016. PMID: 27826850 Free PMC article. Review.
-
Revealing Drivers for Carboxy-S-adenosyl-l-methionine Use by Neomorphic Variants of a DNA Methyltransferase.ACS Chem Biol. 2023 Oct 20;18(10):2224-2232. doi: 10.1021/acschembio.3c00184. Epub 2023 Jun 28. ACS Chem Biol. 2023. PMID: 37379458 Free PMC article.
References
-
- Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin: Landes Bioscience; 2009.
-
- Singer EM, Smith SS. Nucleoprotein assemblies for cellular biomarker detection. Nano Lett. 2006;6:1184–1189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

