tRNA over-expression in breast cancer and functional consequences
- PMID: 19783824
- PMCID: PMC2790902
- DOI: 10.1093/nar/gkp787
tRNA over-expression in breast cancer and functional consequences
Abstract
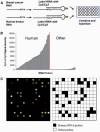
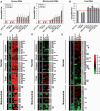
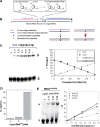
Increased proliferation and elevated levels of protein synthesis are characteristics of transformed and tumor cells. Though components of the translation machinery are often misregulated in cancers, what role tRNA plays in cancer cells has not been explored. We compare genome-wide tRNA expression in cancer-derived versus non-cancer-derived breast cell lines, as well as tRNA expression in breast tumors versus normal breast tissues. In cancer-derived versus non-cancer-derived cell lines, nuclear-encoded tRNAs increase by up to 3-fold and mitochondrial-encoded tRNAs increase by up to 5-fold. In tumors versus normal breast tissues, both nuclear- and mitochondrial-encoded tRNAs increase up to 10-fold. This tRNA over-expression is selective and coordinates with the properties of cognate amino acids. Nuclear- and mitochondrial-encoded tRNAs exhibit distinct expression patterns, indicating that tRNAs can be used as biomarkers for breast cancer. We also performed association analysis for codon usage-tRNA expression for the cell lines. tRNA isoacceptor expression levels are not geared towards optimal translation of house-keeping or cell line specific genes. Instead, tRNA isoacceptor expression levels may favor the translation of cancer-related genes having regulatory roles. Our results suggest a functional consequence of tRNA over-expression in tumor cells. tRNA isoacceptor over-expression may increase the translational efficiency of genes relevant to cancer development and progression.
Figures



References
-
- Bjornsti MA, Houghton PJ. Lost in translation: dysregulation of cap-dependent translation and cancer. Cancer Cell. 2004;5:519–523. - PubMed
-
- Pandolfi PP. Aberrant mRNA translation in cancer pathogenesis: an old concept revisited comes finally of age. Oncogene. 2004;23:3134–3137. - PubMed
-
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. - PubMed
-
- White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

