TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders
- PMID: 19785756
- PMCID: PMC2763876
- DOI: 10.1186/1756-0500-2-197
TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders
Abstract
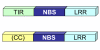
Background: Plant resistance (R) gene products recognize pathogen effector molecules. Many R genes code for proteins containing nucleotide binding site (NBS) and C-terminal leucine-rich repeat (LRR) domains. NBS-LRR proteins can be divided into two groups, TIR-NBS-LRR and non-TIR-NBS-LRR, based on the structure of the N-terminal domain. Although both classes are clearly present in gymnosperms and eudicots, only non-TIR sequences have been found consistently in monocots. Since most studies in monocots have been limited to agriculturally important grasses, it is difficult to draw conclusions. The purpose of our study was to look for evidence of these sequences in additional monocot orders.
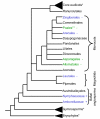
Findings: Using degenerate PCR, we amplified NBS sequences from four monocot species (C. blanda, D. marginata, S. trifasciata, and Spathiphyllum sp.), a gymnosperm (C. revoluta) and a eudicot (C. canephora). We successfully amplified TIR-NBS-LRR sequences from dicot and gymnosperm DNA, but not from monocot DNA. Using databases, we obtained NBS sequences from additional monocots, magnoliids and basal angiosperms. TIR-type sequences were not present in monocot or magnoliid sequences, but were present in the basal angiosperms. Phylogenetic analysis supported a single TIR clade and multiple non-TIR clades.
Conclusion: We were unable to find monocot TIR-NBS-LRR sequences by PCR amplification or database searches. In contrast to previous studies, our results represent five monocot orders (Poales, Zingiberales, Arecales, Asparagales, and Alismatales). Our results establish the presence of TIR-NBS-LRR sequences in basal angiosperms and suggest that although these sequences were present in early land plants, they have been reduced significantly in monocots and magnoliids.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

