Betaalpha-hairpin clamps brace betaalphabeta modules and can make substantive contributions to the stability of TIM barrel proteins
- PMID: 19787060
- PMCID: PMC2747017
- DOI: 10.1371/journal.pone.0007179
Betaalpha-hairpin clamps brace betaalphabeta modules and can make substantive contributions to the stability of TIM barrel proteins
Abstract
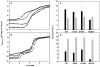
Non-local hydrogen bonding interactions between main chain amide hydrogen atoms and polar side chain acceptors that bracket consecutive betaalpha or alphabeta elements of secondary structure in alphaTS from E. coli, a TIM barrel protein, have previously been found to contribute 4-6 kcal mol(-1) to the stability of the native conformation. Experimental analysis of similar betaalpha-hairpin clamps in a homologous pair of TIM barrel proteins of low sequence identity, IGPS from S. solfataricus and E. coli, reveals that this dramatic enhancement of stability is not unique to alphaTS. A survey of 71 TIM barrel proteins demonstrates a 4-fold symmetry for the placement of betaalpha-hairpin clamps, bracing the fundamental betaalphabeta building block and defining its register in the (betaalpha)(8) motif. The preferred sequences and locations of betaalpha-hairpin clamps will enhance structure prediction algorithms and provide a strategy for engineering stability in TIM barrel proteins.
Conflict of interest statement
Figures
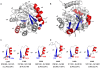



 ), is indicated. The one letter code for the most common amino acids (>15%) in the loop preceding the β-strand in the 71 TIM barrel proteins database is shown. The H-bond network for the β-barrel (
), is indicated. The one letter code for the most common amino acids (>15%) in the loop preceding the β-strand in the 71 TIM barrel proteins database is shown. The H-bond network for the β-barrel ( ), the βα-hairpin clamp interactions between the second residue of an odd-numbered β-strand and the side chain of the residue immediately preceding the subsequent even-numbered β-strand (→), and the MC-MC interactions between the same two residues (− →) are indicated.
), the βα-hairpin clamp interactions between the second residue of an odd-numbered β-strand and the side chain of the residue immediately preceding the subsequent even-numbered β-strand (→), and the MC-MC interactions between the same two residues (− →) are indicated.References
-
- Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. - PubMed
-
- Gerstein M. A structural census of genomes: comparing bacterial, eukaryotic, and archaeal genomes in terms of protein structure. J Mol Biol. 1997;274:562–576. - PubMed
-
- Frenkel ZM, Trifonov EN. Closed loops of TIM barrel protein fold. J Biomol Struct Dyn. 2005;22:643–656. - PubMed
-
- FarzadFard F, Gharaei N, Pezeshk H, Marashi S-A. [beta]-Sheet capping: Signals that initiate and terminate [beta]-sheet formation. Journal of Structural Biology. 2008;161:101–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

