Novel synthetic inhibitors of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase activity that inhibit tumor cell proliferation and are structurally unrelated to existing statins
- PMID: 19787197
- PMCID: PMC2941436
- DOI: 10.3892/ijmm_00000274
Novel synthetic inhibitors of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase activity that inhibit tumor cell proliferation and are structurally unrelated to existing statins
Abstract
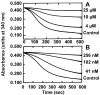
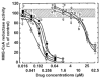
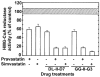
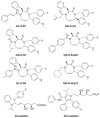
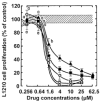
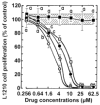
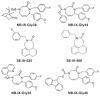
Pilot-scale libraries of eight-membered medium ring lactams (MRLs) and related tricyclic compounds (either seven-membered lactams, thiolactams or amines) were screened for their ability to inhibit the catalytic activity of human recombinant 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in vitro. A dozen of the synthetic compounds mimic the inhibition of purified HMG-CoA reductase activity caused by pravastatin, fluvastatin and sodium salts of lovastatin, mevastatin and simvastatin in this cell-free assay, suggesting direct interaction with the rate-limiting enzyme of cholesterol biosynthesis. Moreover, several MRLs inhibit the metabolic activity of L1210 tumor cells in vitro to a greater degree than fluvastatin, lovastatin, mevastatin and simvastatin, whereas pravastatin is inactive. Although the correlation between the concentration-dependent inhibitions of HMG-CoA reductase activity over 10 min in the cell-free assay and L1210 tumor cell proliferation over 4 days in culture is unclear, some bioactive MRLs elicit interesting combinations of statin-like (IC50: 7.4-8.0 microM) and anti-tumor (IC50: 1.4-2.3 microM) activities. The HMG-CoA reductase-inhibiting activities of pravastatin and an MRL persist in the presence of increasing concentrations of NADPH. But increasing concentrations of HMG-CoA block the HMG-CoA reductase-inhibiting activity of pravastatin without altering that of an MRL, suggesting that MRLs and existing statins may have different mechanisms of enzyme interaction and inhibition. When tested together, suboptimal concentrations of synthetic MRLs and existing statins have additive inhibitory effects on HMG-CoA reductase activity. Preliminary molecular docking studies with MRL-based inhibitors indicate that these ligands fit sterically well into the HMG-CoA reductase statin-binding receptor model and, in contrast to mevastatin, may occupy a narrow channel housing the pyridinium moiety on NADP+.
Figures




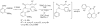
References
-
- Tapiolas DM, Roman M, Fenical W, Stout TJ, Clardy J. Octalactins A and B: cytotoxic eight-membered-ring lactones from a marine bacterium, Streptomyces sp. J Am Chem Soc. 1991;113:4682–4683.
-
- Perchellet JP, Perchellet EM, Newell SW, Freeman JA, Ladesich JB, Jeong Y, Sato N, Buszek KR. Antitumor activity of novel octalactin A analogs in murine leukemia cells in vitro. Anticancer Res. 1998;18:97–106. - PubMed
-
- Buszek KR. First intramolecular benzyne Diels-Alder reaction with an acyclic diene. Unusual effect of diene geometry on the course of the reaction. Tetrahedron Lett. 1995;36:9125–9128.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

