Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex
- PMID: 19789351
- PMCID: PMC2763035
- DOI: 10.1158/0008-5472.CAN-09-0733
Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex
Abstract
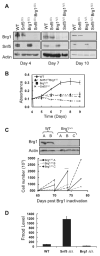
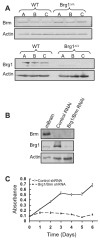
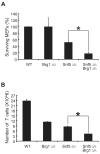
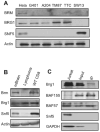
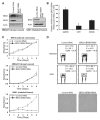
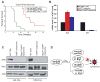
Alterations in chromatin play an important role in oncogenic transformation, although the underlying mechanisms are often poorly understood. The SWI/SNF complex contributes to epigenetic regulation by using the energy of ATP hydrolysis to remodel chromatin and thus regulate transcription of target genes. SNF5, a core subunit of the SWI/SNF complex, is a potent tumor suppressor that is specifically inactivated in several types of human cancer. However, the mechanism by which SNF5 mutation leads to cancer and the role of SNF5 within the SWI/SNF complex remain largely unknown. It has been hypothesized that oncogenesis in the absence of SNF5 occurs due to a loss of function of the SWI/SNF complex. Here, we show, however, distinct effects for inactivation of Snf5 and the ATPase subunit Brg1 in primary cells. Further, using both human cell lines and mouse models, we show that cancer formation in the absence of SNF5 does not result from SWI/SNF inactivation but rather that oncogenesis is dependent on continued presence of BRG1. Collectively, our results show that cancer formation in the absence of SNF5 is dependent on the activity of the residual BRG1-containing SWI/SNF complex. These findings suggest that, much like the concept of oncogene addiction, targeted inhibition of SWI/SNF ATPase activity may be an effective therapeutic approach for aggressive SNF5-deficient human tumors.
Figures
References
-
- Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–9. - PubMed
-
- Versteege I, Sevenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. - PubMed
-
- Kohashi K, Oda Y, Yamamoto H, et al. SMARCB1/INI1 protein expression in round cell soft tissue sarcomas associated with chromosomal translocations involving EWS: a special reference to SMARCB1/INI1 negative variant extraskeletal myxoid chondrosarcoma. Am J Surg Pathol. 2008;32:1168–74. - PubMed
-
- Modena P, Lualdi E, Facchinetti F, et al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res. 2005;65:4012–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

